I denne pilotstudien har vi testet effektiviteten av ulike metoder for å kartlegge og overvåke fremmede marine arter i tre havner i Rogaland fylke. Vi har brukt metoder som er anbefalt i HELCOM protokoller som er utviklet for Østersjøen, men har også inkludert strandsøk, videoundersøkelser, trekantskrape og en mer omfattende bruk av genetiske metoder. Materiale fra dyreplanktonprøver og fra begroingspaneler, samt miljø-DNA prøver fra sediment og vannprøver er analysert med metastrekkoding. Det ble totalt funnet 33 fremmede arter i denne studien hvorav syv arter var dørstokkarter for Norge. De konvensjonelle metodene registrerte 19 fremmede arter og metodene RCS i båthavner, begroingspanel og strandsøk var de mest effektive metodene. Genetiske metoder registrerte 26 fremmede arter og her var analyser av vannprøver og avskrapt materiale fra begroingspanel de mest effektive. Vi anbefaler en kombinasjon av konvensjonelle metoder og DNA-analyser for et nasjonalt overvåkningsprogram. Fangst av mobil fauna i teiner registrerte ingen fremmede arter i pilotområdet, men vi anbefaler likevel denne metoden da den er den beste metoden for å fange fremmede krabber og fisk.
Monitoring marine alien species in Norway
— A pilot study for implementing a national program
Rapportserie:
Rapport fra havforskningen 2024-1
ISSN: 1893-4536
Publisert: 12.02.2024
Oppdatert: 24.11.2025
Prosjektnr: 15635-03
Oppdragsgiver(e): Miljødirektoratet
Referanse: 2021/5221
Forskningsgruppe(r):
Bentiske ressurser og prosesser
Program:
Kystøkosystemer
Approved by:
Research Director(s):
Geir Huse
Program leader(s):
Jan Atle Knutsen
English summary
Forord
The Institute of Marine Research was given an assignment from the Norwegian Environment Agency (NEA) in 2021 to compile data on existing mapping and monitoring of alien marine species in Norway and to assess the vectors for introductions into Norwegian waters (Husa mfl. 2022: Alien Marine species in Norway - Mapping, monitoring and assessment of vectors for introductions). Moreover, we suggested methods for a national program for mapping and monitoring of marine alien species (Husa mfl. 2002: Metodikk for kartlegging og overvåkning av fremmede marine arter i Norge - Forslag til nasjonalt program). The third part of the assignment was to test suggested methods in some Norwegian ports and the presented report is the third and last to complete this work. Based on the experience from the testing of methods a detailed national program for monitoring of alien species in ports and marinas in Norway has been delivered NEA with associated budgets (not presented here).
Sammendrag
In this pilot study we tested the efficiency of different methods to detect alien marine species in three different ports in Rogaland County. The methods used followed by far the recommendations in the HELCOM protocol, which is developed for the Baltic Sea, but also included beach survey, video survey, dredging and extensive use of DNA-methods. Material from zooplankton samples, scrapings from settlement panels as well as e-DNA samples from filtrated water and sediments were subject to meta-barcoding. A total of 33 alien species were recorded in this study, of which seven were door knockers to the Norwegian coast. The conventional methods recorded 19 alien species, and the methods RCS in marinas, settlement panels and beach surveys had the highest detection rate. DNA-methods detected 26 alien species, and scrapings from settlement panels and water samples were the most efficient methods. We recommend a combination of conventional and DNA-methods to be used in a national monitoring program. Although sampling of mobile fauna yielded no alien species in the pilot area we still recommend to use this method, being the best method for catching alien crabs and fishes.
1 - Introduction
Marine alien species have an increasing focus globally. A recent review listed 874 non-indigenous marine species (NIMS) being introduced to Europe since 1970 (Zenetos et al. 2022). In Europe, several initiatives have been taken to decrease the introduction of alien species and thereby the impact on marine biodiversity. By the implementation of the Ballast Water Management Convention in 2004 the risk for introducing species in ballast water has diminished considerably. However, species introduced for aquaculture purposes and their associated hitchhikers are still an issue in many European countries. In the Mediterranean, numerous alien species are migrating through the Suez Canal due to ocean warming and continuously widening of the canal. In Norway, the main vector for introduction of marine alien species is biofouling on vessel and floating debris (Husa et al. 2022). Due to its relatively cold climate, the Norwegian coast still holds a low number of marine alien species, but several species have arrived in the recent decades. A total of 43 species have been established and another four species are recorded but not with documented reproducing populations. Most of the species are recorded from the southern and mid part of Norway, while only a few are present in the north (Husa et al. 2022). To understand the dispersal of alien species and the extent of their impact on native communities, mapping and monitoring their distribution is essential. National monitoring programs can provide an early warning of troublesome newcomers and important information can be used to initiate necessary mitigation measures. In Norway there are existing monitoring programs on red king crab, snow crab (Husa et al. 2022), Pacific oysters (Mortensen et al. 2017) and pink salmon (Berntsen et al. 2020). An initiative with targeted mapping of the newly arrived ascidian Didemnum vexillum using a combination of eDNA and visual monitoring has been conducted since 2021 (Fossøy et al. 2022, Fossøy et al. 2023). Mapping of alien biota in marinas from the Swedish border to the county of Møre & Romsdal has been conducted yearly from 2010 to 2023. In this project, a small part of the coast has been mapped for one summer week every year (Husa et al. 2012 ab, Husa et al. 2013, Husa et al. in prep). Two recent reports issued by the Norwegian Environment Agency has already discussed existing literature and the potential for using DNA-based methods for monitoring alien and doorknocker species (Ekrem et al. 2023) and marine biodiversity in general (Dunshea et al. 2021). Several international methods and protocols for how to search for marine alien species in the marine environment already exists, such as the CRIMP protocol developed for Australia (Hewitt & Martin 2001), the Rapid Coastal Survey (RCS) used in many countries (Minchin 2007), the SERC-protocol for NIS in the US (Marrafini et al. 2017) and the HELCOM protocol which was developed for the Baltic Sea. CRIMP (Centre for Research on Introduced Marine Pest) have tested out and published revised protocols for baseline port surveys for introduced Marine species in Australia (Hewitt & martin 2001). This is an extensive protocol but also gives good baseline data for the ports. The method involves vast use of divers, but also opens for alternative sampling by grab/sledge and video surveys. The RCS method (Rapid Coastal Survey), also called RAS (Rapid Assessment Survey), is basically a quick investigation of all fauna and flora growing on floating docks and ropes in marinas. This method has proven efficient to detect alien species at low costs and has also been used in Norway (Husa et al. 2012 a, b, Husa et al. 2013). The SERC-protocol ( Smithsonian Environmental Research Center) has been developed and tested out for the US and focuses on deployment of settlement panel to detect alien marine species. The method easily captures sessile organisms but is less efficient detecting species associated with other substrate than hard bottom (Marrafini et al. 2017). The HELCOM protocols are building upon the CRIMP protocol but are using more methods that can be performed from land to make the monitoring less costly than the with the use of divers. HELCOM provides a series of detailed protocols for sampling of phytoplankton and zooplankton, infauna in sediments, mobile and sessile fauna, settlement panels, investigations of marinas, and genetic methods (HELCOM 2013).
Denmark has developed a national program for monitoring of marine alien species based on the HELCOM protocol, but also including environmental DNA (eDNA) and DNA metabarcoding (Andersen et al. 2016, 2017, 2018, 2022). Finland has developed a similar approach (HELCOM 2013). The use of eDNA from water-samples, sediment samples, scraping and plankton samples to detect marine alien species has been tested in several studies (Taberlet et al. 2012, Ardura et al. 2015, Forsström & Vasemägi 2016, Miralles et al. 2016, Ardura & Zaiko 2018, Muñoz-Colmenero et al. 2018, Fernandez et al. 2021) and has proven useful to detect rare alien species but also generates a high amount of data which is not necessarily useful for management (Sepulveda mfl. 2020).
As the HELCOM method is suggested for many European countries the method is probably also the most suitable protocol for a Norwegian monitoring programme. The aim of this study is to test the efficiency of conventional methods (actual sampling of organisms), as well as e-DNA and DNA metabarcoding for monitoring marine alien species at the Norwegian coast. Based on the results we will recommend methods suitable in a Norwegian National program for monitoring of marine alien species.
2 - Material and methods
2.1 - Target species
A list for marine alien species for Norwegian waters was created for macroalgae, marine invertebrates and fish based on lists from the Horizon scanning and risk assessment performed by the Norwegian Biodiversity Information Centre in 2022-2023. This target list contains alien species that are established in Norwegian waters, species that are observed here without documented viable populations and species that are considered door knockers. List of target species is to be found in Appendix Table 1, 2, 3. For each target species on the list, information from genetic databases was gathered to check if sufficient information were present.
2.1.1 - ID material
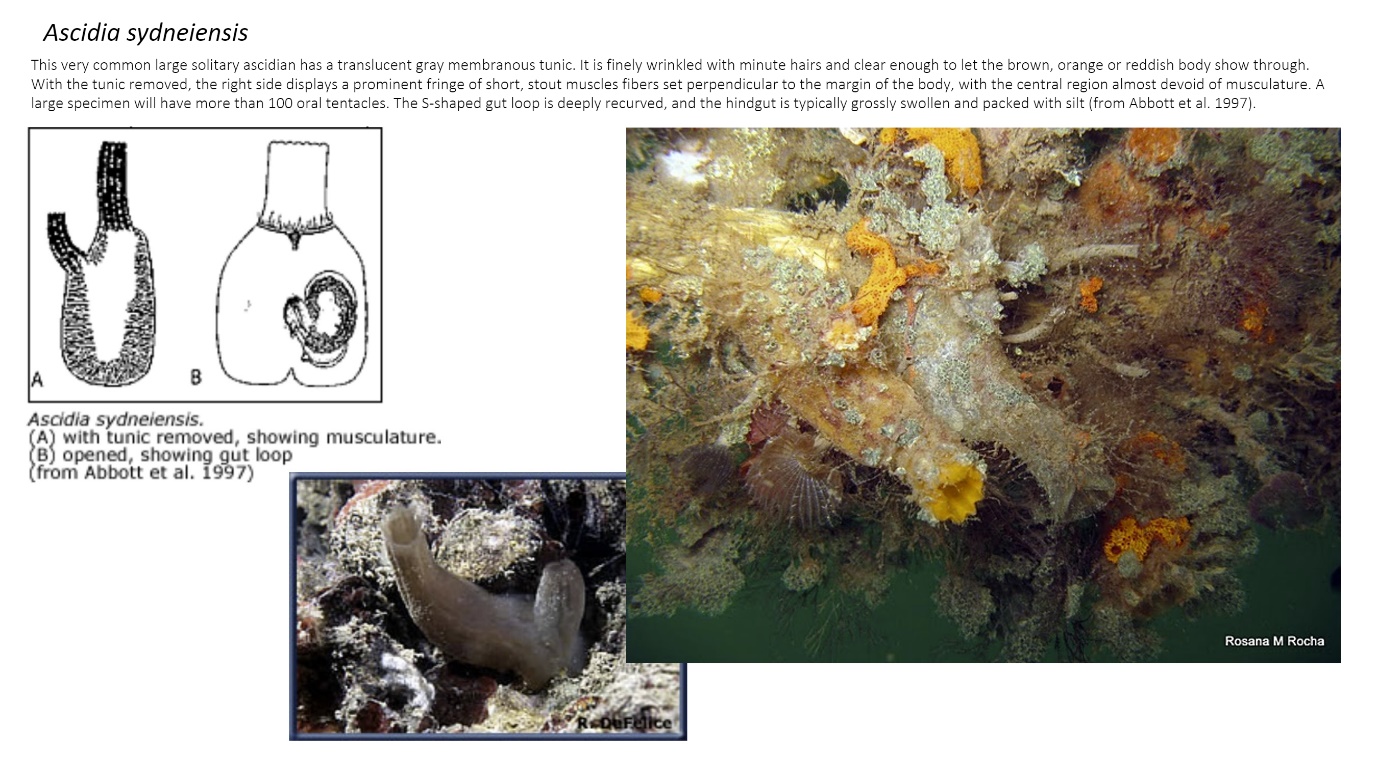
For each target species an identification card was created with photos and drawings of morphology and characteristics, relevant information on how to identify the species and references to taxonomic literature. The ID cards were gathered in collections related to taxonomic groups and where they were going to be used e. g. jelly plankton (field) zoo plankton (laboratory). Figure 1 shows and example of an ID card for door knocker Ascidia sydneiensis .
2.2 - Study area
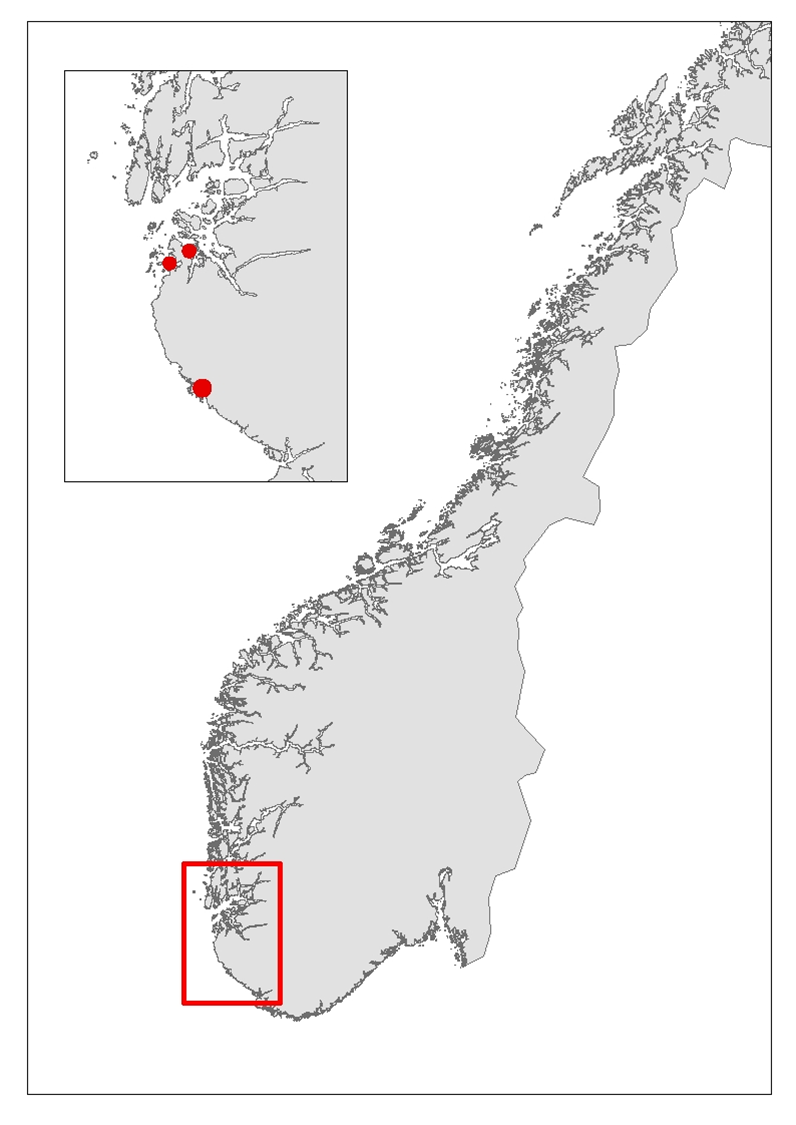
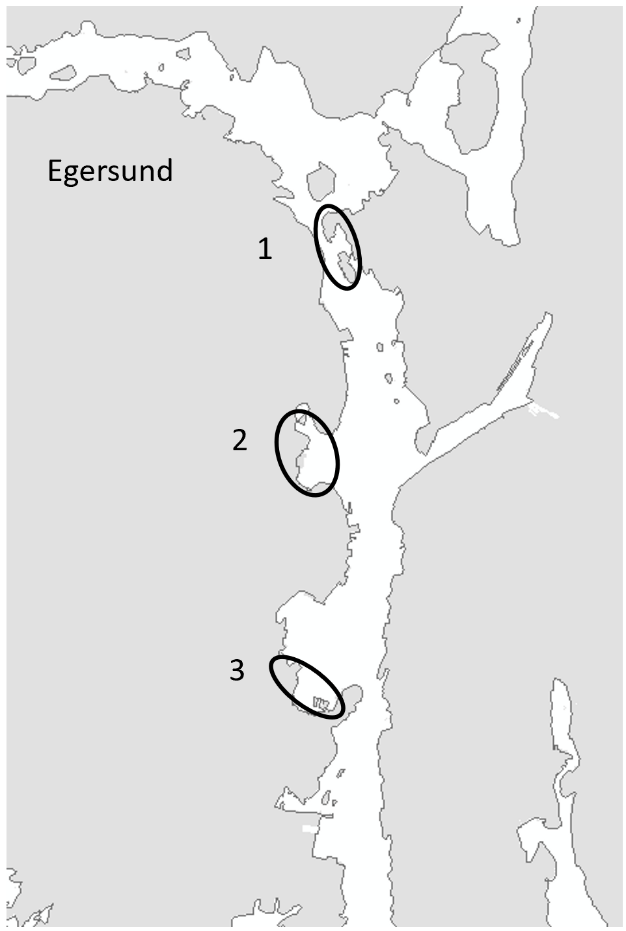
Three ports, Stavanger, Tananger and Egersund in Rogaland County (Figure 2) were chosen based on high frequency of foreign ships arrivals and proximity to each other to ease fieldwork and reduce travel costs. Rogaland County is the one of the regions in Norway with the highest annual number of foreign ship arrivals, which increases the risk for introduction of marine species through biofouling of the ship hull (Husa et al. 2022). Rogaland is also one of the regions with the highest number of established marine alien species (Sandvik et al. 2019, Husa et al. 2022).
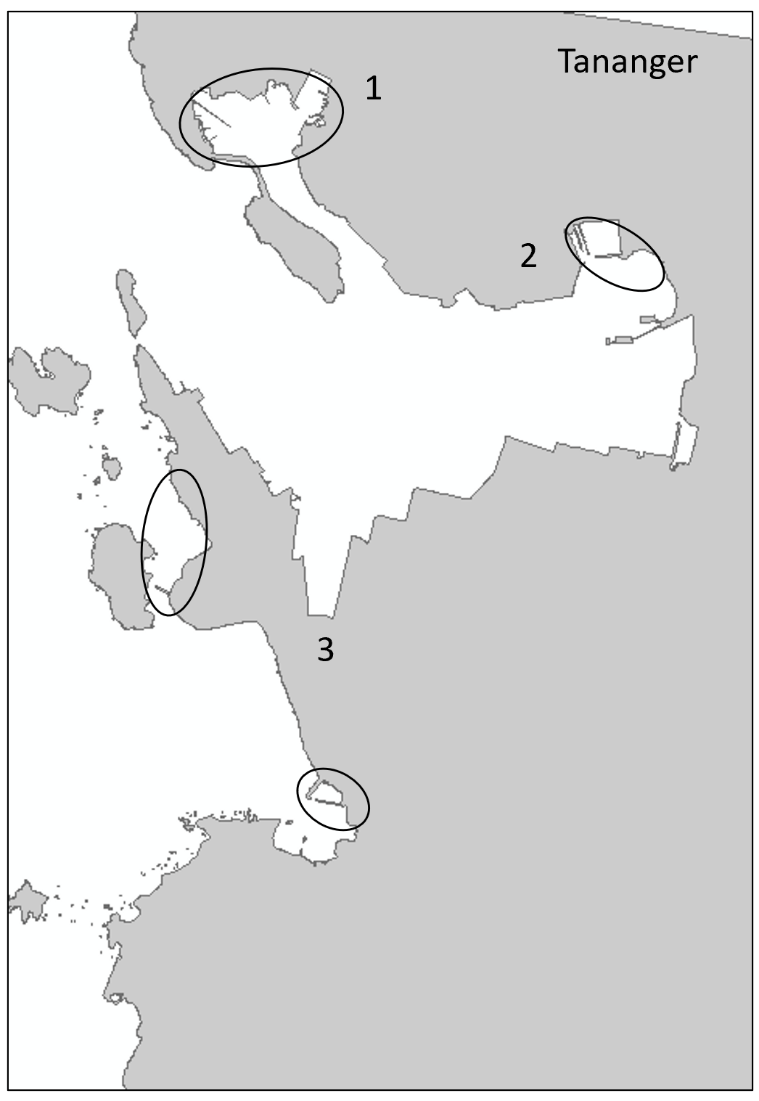
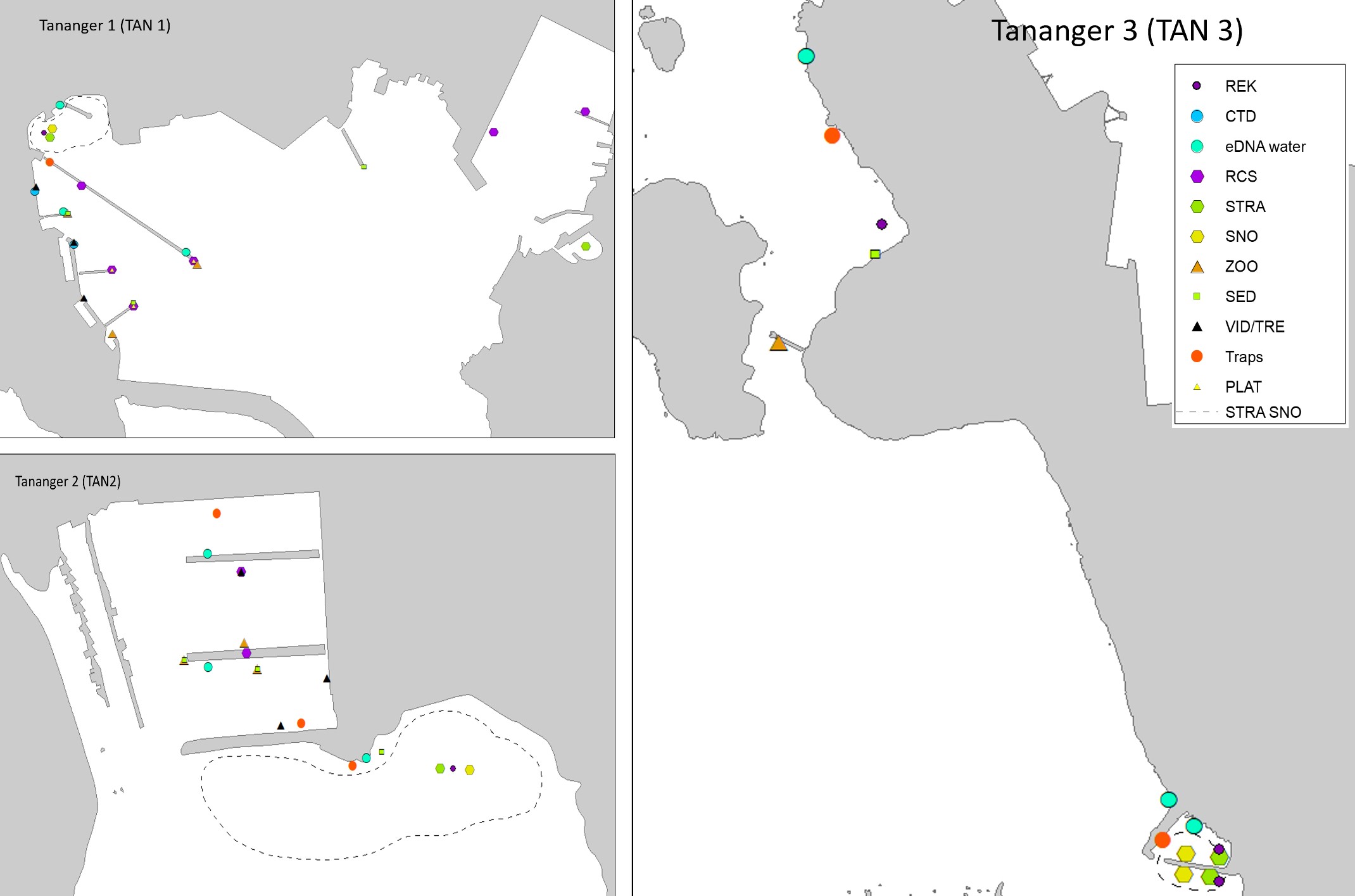
Tananger is an industrial port situated in a bay on the west coast of Stavanger. The port receives approximately 700 foreign arrivals annually, mostly cargo vessels but also tankers and special crafts. In addition, there is direct ferries from Denmark every second day. Most of the vessels comes from ports in the Netherlands, United Kingdom, Germany, Sweden, and Denmark (Husa et al. 2022 a). The bay is 42 meters deep at the deepest and has shallow sandy areas in the inner part and on the west side of the industrial area. The area has three smaller marinas. The bay has fully marine conditions with low freshwater impact and good exchange of water at the inlet. Overview of stations examined in this port is given in Figure 3. Station three (TAN 3) is divided into two areas to cover sufficient habitats.
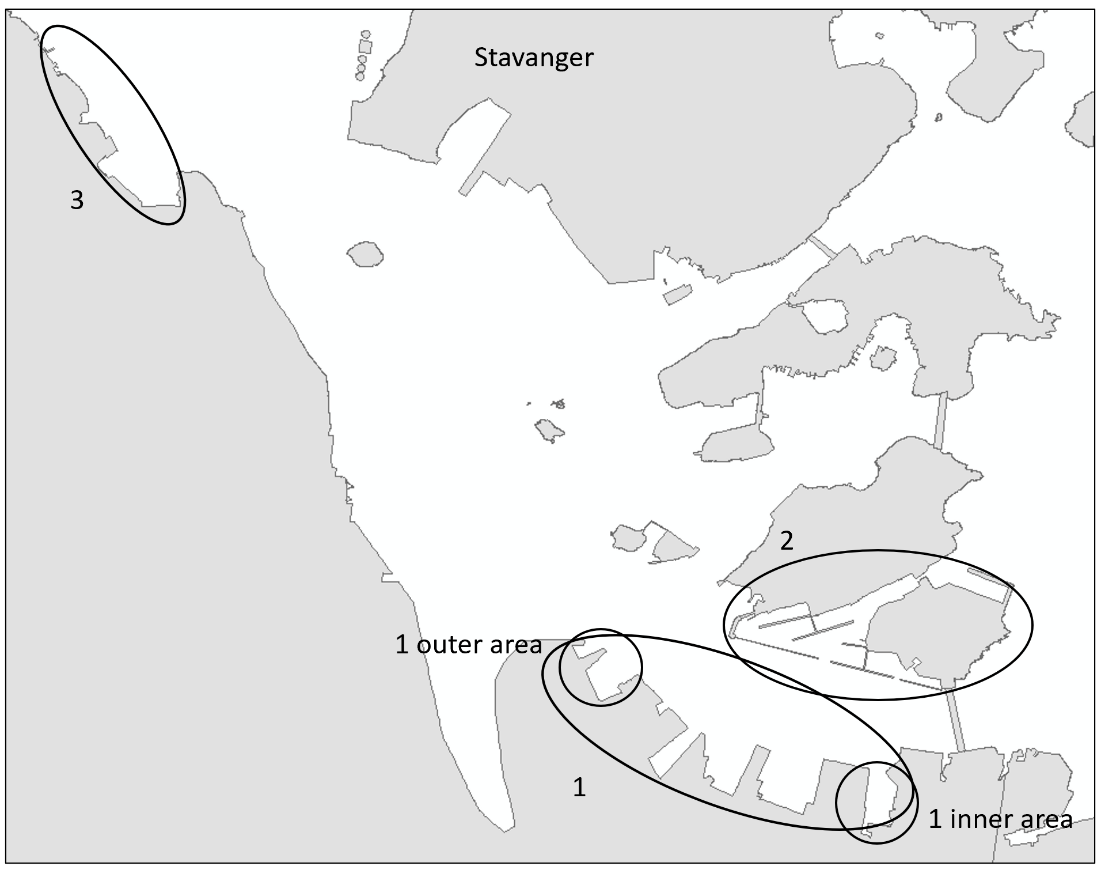
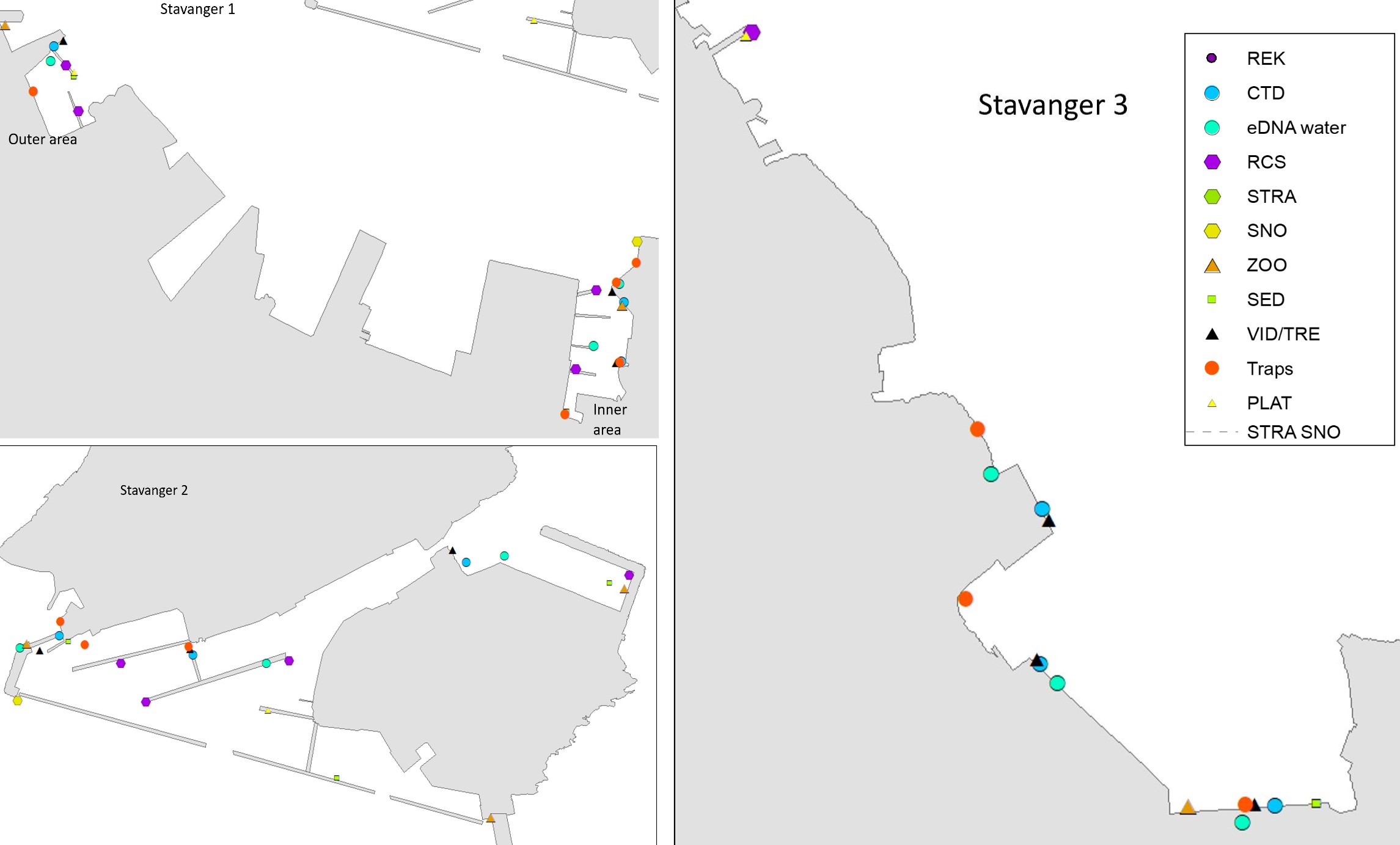
Stavanger port authority covers a larger area with several keys in Stavanger city and additional keys and shipping wharfs in the vicinity. There are several large marinas in the area. The port receives approximately 250 arrivals from foreign ports annually, divided by cargo and special crafts, and some tankers. Most of the foreign arrivals are from ports in Netherlands, United Kingdom, Germany, Sweden, Denmark, and the Baltic Sea (Husa et al. 2022). Stavanger port is a busy cruise port and receives frequent visits from pleasure- and fishing boats. The area has a maximum depth of 56 meters and are fully marine with good water exchange. Overview of stations examined in this port is given in Figure 4.
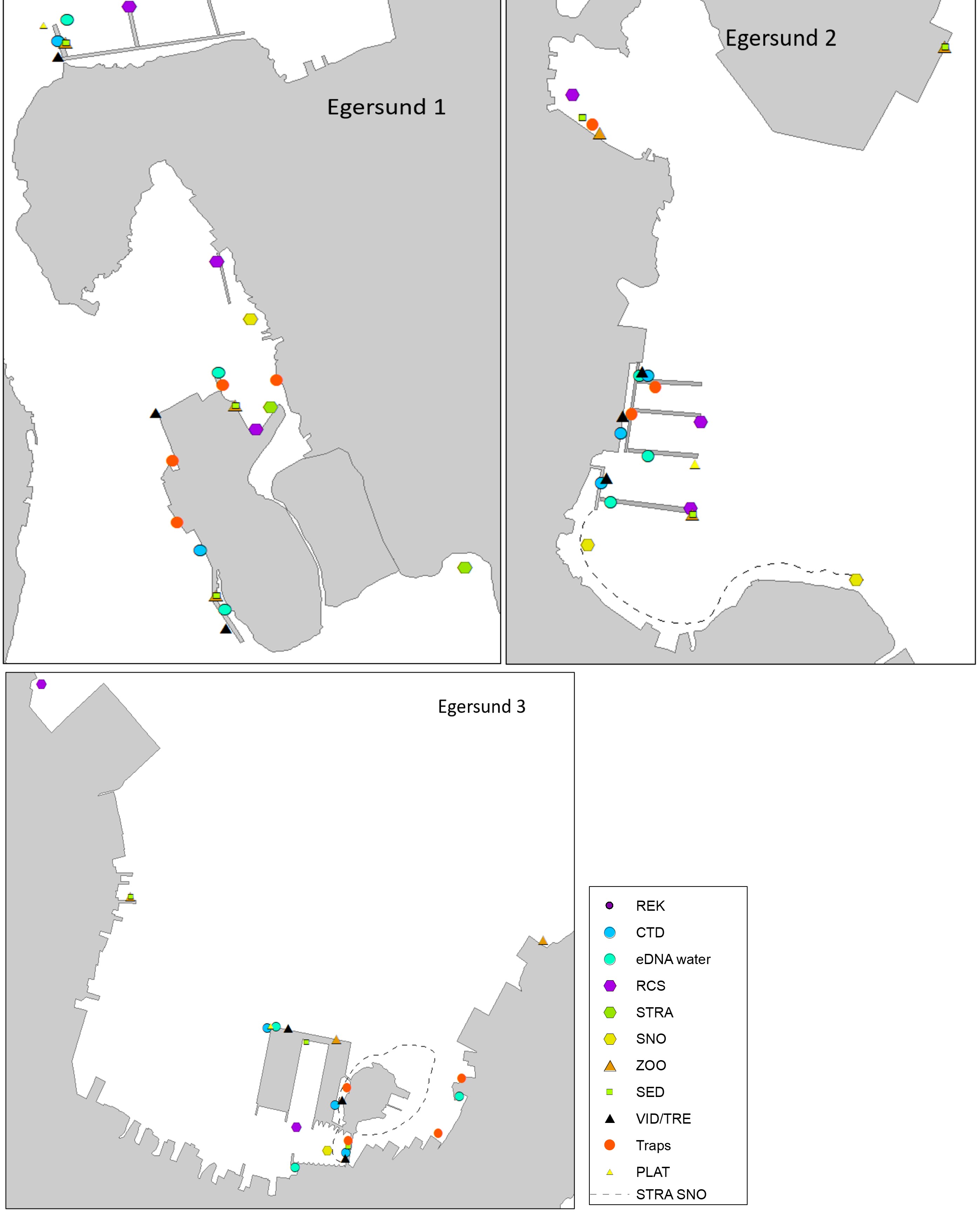
Egersund port is a wide area situated in a current rich sound. Most of the sound has depths shallower than 10 meters and inner areas are strongly influenced by freshwater. The port has various keys, marinas, and some industry keys. The port receives approximately 150 foreign arrivals a year, mostly cargo vessels from Germany, the Netherlands, United Kingdom, Denmark, and the Baltic Sea. The port is also a busy port for fishing vessels, mostly local but also some arrivals from Denmark, Faroe Islands and United Kingdom. The tidal amplitude in Egersund is zero, so high and low water is only driven by meteorological forces. Overview of stations examined in this port is given in Figure 5.
2.2.1 - Selection of sampling sites
The HELCOM protocol recommends sampling of minimum three sites within each port with a higher number for large ports to include all types of habitats. We selected three sites in each port based on the following criteria: closeness to human marine activity, presence of at least one floating dock or pier preferably a marina, presence of both hard and soft natural substrate with an accessible shoreline. The study sites in each port are presented in Figures 3-5. An overview of methods used at each station is given in Figures 6 to 8.
Tananger port
2.3 - Sampling methods
Both HELCOM and CRIMP recommend sampling of pathogens in seawater and sampling of phytoplankton. We did not include this in our sampling program since both pathogens and phytoplankton in seawater is already included in regularly monitoring programs by Norwegian authorities (https://algestatus.hi.no/, https://lovdata.no/dokument/SF/forskrift/2006-12-15-1446).
2.3.1 - Salinity and temperature
At each site salinity, temperature and turbidity were measured with a RBR logger (RBRconcerto) from the surface to ½ meter above the bottom. Such measurements are recommended in both the HELCOM and the CRIMP protocol but will only give a picture of the given conditions during the sampling day. At some sites the water was so shallow that it is only point measurement.
2.3.2 - Zooplankton
At each site a minimum of three vertical hauls from the bottom and up were performed with plankton net 180 µm. Additional horizontal hauls were taken if there was little material in the nets. At some stations it was so shallow that only horizontal hauls were possible. Jelly plankton were separated from the sample and identified in the field or brought alive back to the field lab for identification. The remaining material from the hauls at each site were pooled and divided in two vials and preserved with 96% ethanol. The fauna in one vial from each site were identified in the lab and the other vial were object to DNA-metabarcoding.
2.3.3 - Infauna in sediments
Grab samples were taken with van Veen grab (250 cm²) from three places with minimum 15 meters distance at each station. Each place was sampled by at least one grab to ensure sufficient material (1/3 full grab or minimum 10 cm filled). The quality of the sediment was classified by smell, colour, and texture. The material was rinsed through a 1 mm sieve and preserved in 4 % formaldehyde and 1 tablespoon of Borax and later transferred to 96 % ethanol. All fauna was identified in the lab to lowest taxonomic level following nomenclature in Worms. Samples for DNA-metabarcoding of sediment were taken from the surface layer of multiple grabs and/or different places in each grab and pooled together as one sample from each station.
2.3.4 - Settlement plates
Three rubbed plastic plates (Styren akrylonitril) 15x15 cm were attached to a rope at 1-, 3- and 5-meters depth with a weight in the deep end of the rope. Three ropes (nine plates) were deployed at three different floating docks in each area at the end of June 2022 and collected again in November 2022. The plates were brought back to lab in fresh seawater in a cooling box and photographed at each side. Large organisms were identified and removed from the panels. The remaining material was divided in two where one half of the plates on both sides were examined in a stereoscope and identified to lowest possible taxonomic level. The other half of the plates were scraped off and preserved in ethanol for DNA-metabarcoding.
2.3.5 - e-DNA water samples
At each site, between 3.5 and 8 litres of surface water were collected and filtered in situ using a Bürkle Vampire handheld pump and 0.8 µm filters (Nature Metrics). Care was taken to avoid contamination by using single-use clean buckets and rope, and ready-made kits for each site with gloves, tubing, syringes with buffer, filters, and caps. Samples were conserved with ATL-buffer after filtration and stored in room temperature until genetic analyses at The Centre for Biodiversity Genetics (NINAGEN) in Trondheim.
2.3.6 - Crab pots and minnow traps
Three square crab pots (60x45 cm) with an escape opening of 6 cm and five aluminium minnow traps (24 cm diameter) with an escape opening of five cm (Figure 9) were baited with boiled shrimp and deployed close to land at depths between one and five meters. The traps and pots had a soak time between 12 and 24 hours. Organisms collected in the traps were identified in field.

Figure 9. Minnow traps baited with shrimps used for capturing mobile fauna at shallow depth.
2.3.7 - Dredging and video transects
Three video-transects of 10 meters along floating docks or piers were conducted approximately 50-70 cm above the seafloor at each station by using a video rigg with a mounted action camera (GoPro HERO 9), a navigation camera and a light. Material in the transect were collected with a handheld dredge at the same distance. Larger organisms from the dredge were identified in the field while specimens that needed closer examination were brought back to the field lab in fresh seawater. Video records were analysed for presence of alien species.
2.3.8 - Sampling of sessile organisms in marinas - Rapid Coastal survey (RCS)
This investigation was performed in marinas at each station when applicable. At some sites there were not true marinas but merely smaller boat harbours with a few jetties. Following HELCOM procedures the marina was divided into an outer and inner area. Three floating docks in each area were studied by taking vertical scraping samples at 3 places on each dock. Additionally, hanging ropes were elevated and samples were taken from different depths on the rope. A visual inspection of one dock in each area was also performed to look for alien species. Easily recognizable species, native and non-native, were sorted out in field and noted. The remaining material was brought back fresh in seawater to the field lab and identified to lowest taxonomic level. Species that were not identified in the field lab were fixated in 96% ethanol and sent to experts or genetic analysis.
2.3.9 - Sampling of organisms on beaches-Beach survey
At sites which had a beach stretch consisting of sand, gravel or rocks, a 100 meter long transect were subject to investigation in the following manner: a) collection of species by snorkeling, b) collection of species by walking/wading along the shore, c) three fucoid and three seagrass plants (if present) were sampled by placing a plastic bag over the plant to collect associated fauna, d). a shrimp collection net (Figure 10) was used to collect mobile fauna at the bottom of the survey stretch. When possible, stones were lifted to bring animals out in the open. All species were brought back to the field lab for identification to lowest possible taxonomic level.

2.3.10 - Patogenes in mollusks
During sampling on beaches and on floating docks, samples of oysters were collected to examine those for shellfish pathogens. In oysters there were a specific focus on blister worms. Oysters were opened and examined for blisters, whenever those were present, they were broken and examined for worms.
2.3.11 - Genetic analyses
2.3.11.1 - DNA-extraction
Filtered eDNA water samples, zooplankton, sediments, and settlement plates were subject to DNA-metabarcoding for species identification. The extraction of DNA from the water filters was initiated by adding 130 μL proteinase-K (diluted 1:10) to the filters before incubation at 56°C overnight. DNA was extracted using a combination of NucleoSpin Plant II (Machery-Nagel) spin columns and Blood & Tissue buffers (Qiagen). DNA was eluted in 200 μL pre-heated AE-buffer (Qiagen) and thereafter re-eluted for maximizing the DNA-output.
The sediment, plankton and scrape-off from settlement plates arrived at NINAGEN in 50 mL tubes. The tubes were added 20 g Matrix D (MP Biomedicals), 8 ceramic spheres (MP Biomedicals) and 0.5 mL Matrix A garnet (MP Biomedicals) and filled up to 35 mL with 96 % ethanol. The tubes were then homogenized in a FastPrep-96 homogenizer (MP Biomedicals) for 1 min at 1600 rpm, followed by homogenizing in a FastPrep-24 homogenizer (MP Biomedicals) for 40 sec at 4.0 m/s. Subsamples of 500 µl from each sample where dried in a heating cabinet to evaporate the EtOH. When dried, 540 µl ATL-buffer (Qiagen) and 60 µl Proteinase-K (Merck) were added and the samples were incubated overnight at 56 degrees. DNA was then extracted using a MagMax DNA Ultra 2.0 Kit (Thermo Fisher) in a KingFisher Apex extraction robot (Thermo Fisher). DNA was eluted in 250 µl AE-buffer (Qiagen).
2.3.11.2 - DNA metabarcoding
We amplified four different genetic markers across the different sample types (Table 1). We specifically selected markers targeting algae (TAReuk), invertebrates (Leray and modBF) and fish/vertebrates (MiFish), which will provide information on different taxonomic groups. The amplifications were based on the two-step Illumina (San Diego, CA, USA) 16S protocol (Anonymous, 2013). The first PCR reactions had a final volume of 25 μL containing 2.5 μL DNA template (each template was diluted to have 1-10 ng/µL of DNA depending on the marker), 12.5 μL 2x KAPA HiFi HotStart ReadyMix (Roche, Basel, Switzerland), forward primer and revers primer. All PCRs included negative control reactions (no DNA template). The PCR conditions used were as determined by the reference (Table 1) for each marker, with some modification in number of PCR cycles. In the second PCR step, we dual-indexed Illumina-tailed amplicons, using IDT for Illumina DNA/RNA UD indexes (Illumina) under PCR conditions with a heated lid, 95 °C for 3 min, followed by a total of 8 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s, and a final extension at 72 °C for 5 min. The second-step PCRs had a final volume of 50 µL containing 5 µL of the first-step PCR product, 25 µL 2x KAPA HiFi HotStart ReadyMix, 10 µL of forward index, 5 µL of reverse index and 10 µL molecular grade H2O. We visualised the PCR products on a Tape Station (Agilent 4200, Agilent, Santa Clara, CA, USA) to check the amplification success and cleaned them with magnetic beads (MAGBIND TotalPure NGS, Omega Bio-Tek Inc., Norcross, GA, USA) after each PCR step. In the end, we normalized the indexed amplicons based on values from the Tape Station and pooled them into ready libraries. The libraries were sequenced using the Illumina NovaSeq platform at the Norwegian Sequencing Centre (NSC) in Oslo.
Table 1. A list of genetic markers used for species identification with details on target group and sample type analysed as part of this study.
|
Genetic marker |
Forward/reverse primer |
Target group |
Sample types analysed |
Reference |
||
|
Leray |
mlCOIintF2 jgHCO2198_2 |
Invertebrates |
Zooplankton, sediments, settlement plates and water |
Leray et al. (2013) Majaneva unpubl. |
||
|
mod_BF |
BF3_mod BR2_mod |
Invertebrates |
Settlements plates |
Elbrecht et al. (2019) Jensen mfl. (2021) |
||
|
MiFish |
|
Fish |
Water |
Miya et al. (2015) |
||
|
TAReuk |
TAReuk454FWD1 TAReukREV3r |
Algae |
Water and settlement plates |
Stoeck et al. (2010) |
2.3.11.3 - Bioinformatic analyses
The data was demultiplexed and adapters were removed on the Illumina NovaSeq platform. Primers were removed from both the 5’ and 3’ ends of reads using cutadapt v.3.7 (Martin 2011) with a minimum length match of 17 bp and 0.15 expected errors in the primer region. Quality filtering, error correction, merging, mapping and chimera detection in the sequence data was conducted using DADA2 v 1.9 package in R (Callahan et al. 2016). Reads with ambiguous bases, >2 expected errors in the forward and reverse directions, or length <50 bp after truncation at the first instance of a base with a quality score <10, were removed from the dataset. Error rate models with enforced monotonicity were estimated per marker for both forward and reverse sequences. After correction, reads were merged with a minimum overlap of 30 bp. Amplicon sequence variants (ASVs) were inferred for each sample: marker combination and chimeric sequence variants were assessed on a per-sample basis, as chimeric events occur at the individual PCR level. If a sequence variant was flagged as chimeric in more than 90% of the samples it occurred in, it was removed. ASVs were then subjected to megablast searches against the NCBI nucleotide non-redundant database. Only ASVs with a >97% identity and >80% coverage best match to a known reference sequence were retained for analysis. ASV identification confidence was categorized as "moderate” (97-97.9%), “good” (98-99.9%), or “very good” (100%) based on the percent similarity of the best BLAST match.
3 - Results
3.1 - Salinity and temperature
Single measurements of salinity and temperature give us of course no indication of the normal situation in the area but can give us information on the situation in the different areas at the time of the sampling.
Stations in the Stavanger port (STA 1, 2, 3) had stable salinities and temperatures in the water column. Salinities ranged from 28.7 psu at the surface to 29.9 psu at the bottom. Temperatures were ranging from 18.5 o C in the surface water to 17.2 o C at the bottom.
Stations in the Tananger port (TAN 1, 2, 3) were the most marine stations in the study with salinities between 31.6 and 32.2 psu and temperatures between 18.4 o C in the surface and 15.4 o C at 4 meters depth.
Stations in the Egersund port (EG 1, 2, 3) were strongly influenced by freshwater runoff from the river with a shallow brackish surface layer. The EG 1 station was closest to the river and had mean salinities at the surface of 5.9 psu, sharply rising to 27.4 psu at three meters depth. The two stations EG 2 and EG 3 were further away from the river but still had low salinities of 12.9 and 13.7 psu in the surface layer, but sharply rising to 25-26 psu at 3-4 meters depth. Temperatures were more stable with slightly colder surface temperatures of 17.5 o C in the surface and 18.8 o C at 3-4 meters (Detailed CTD data for each station are given in Appendix B).
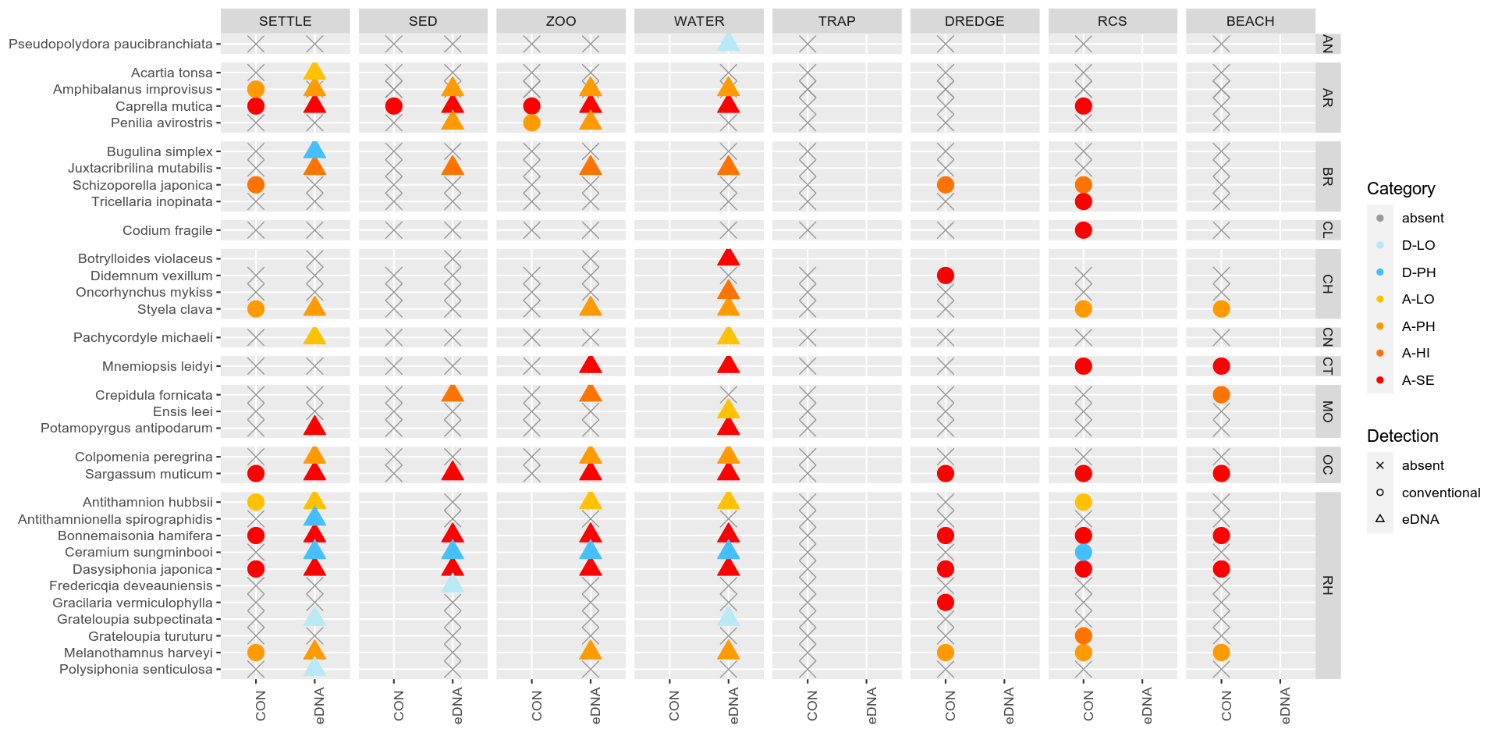
3.2 - Marine species recorded by collection method.
3.2.1 - Zooplankton samples
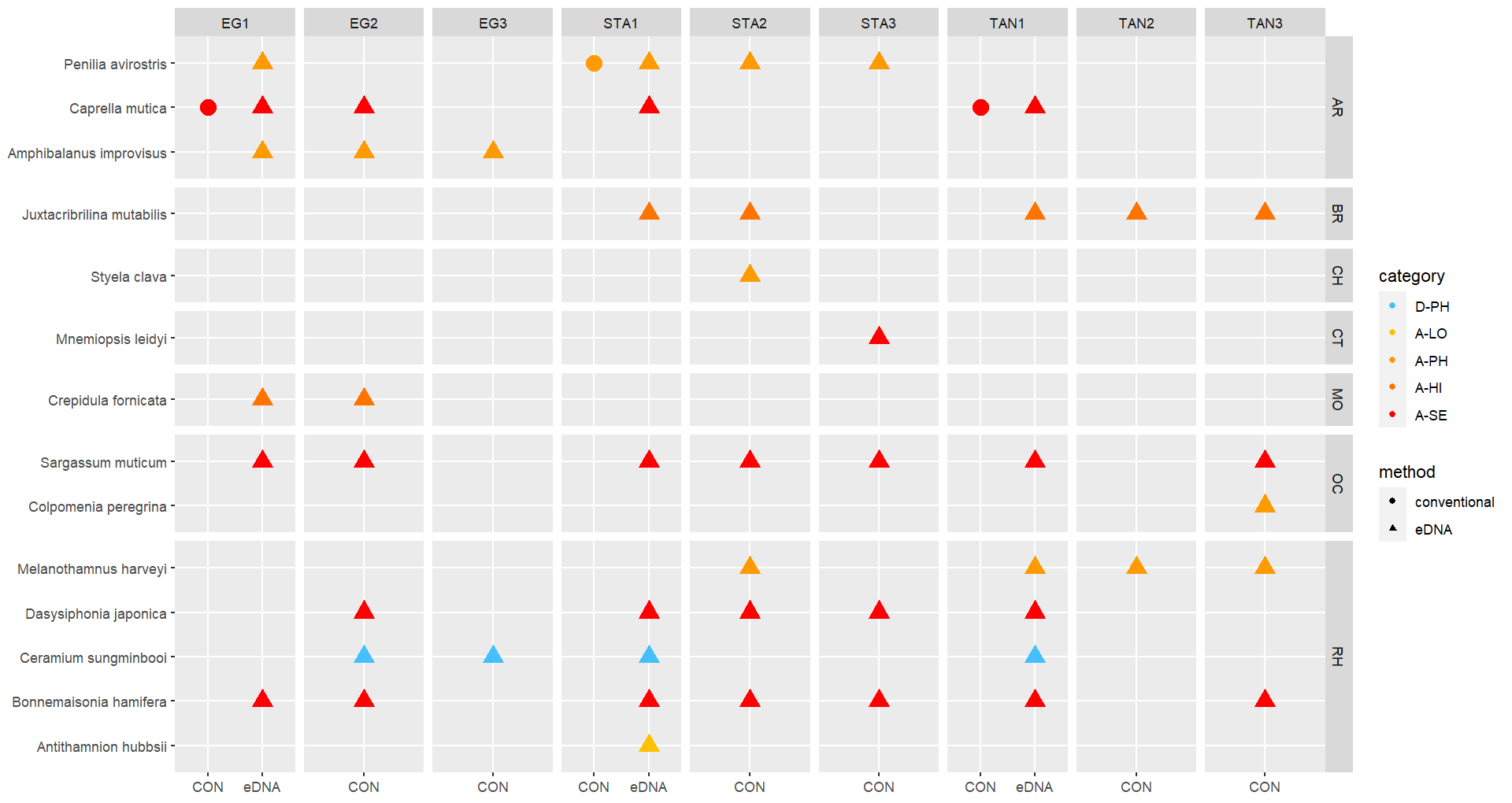
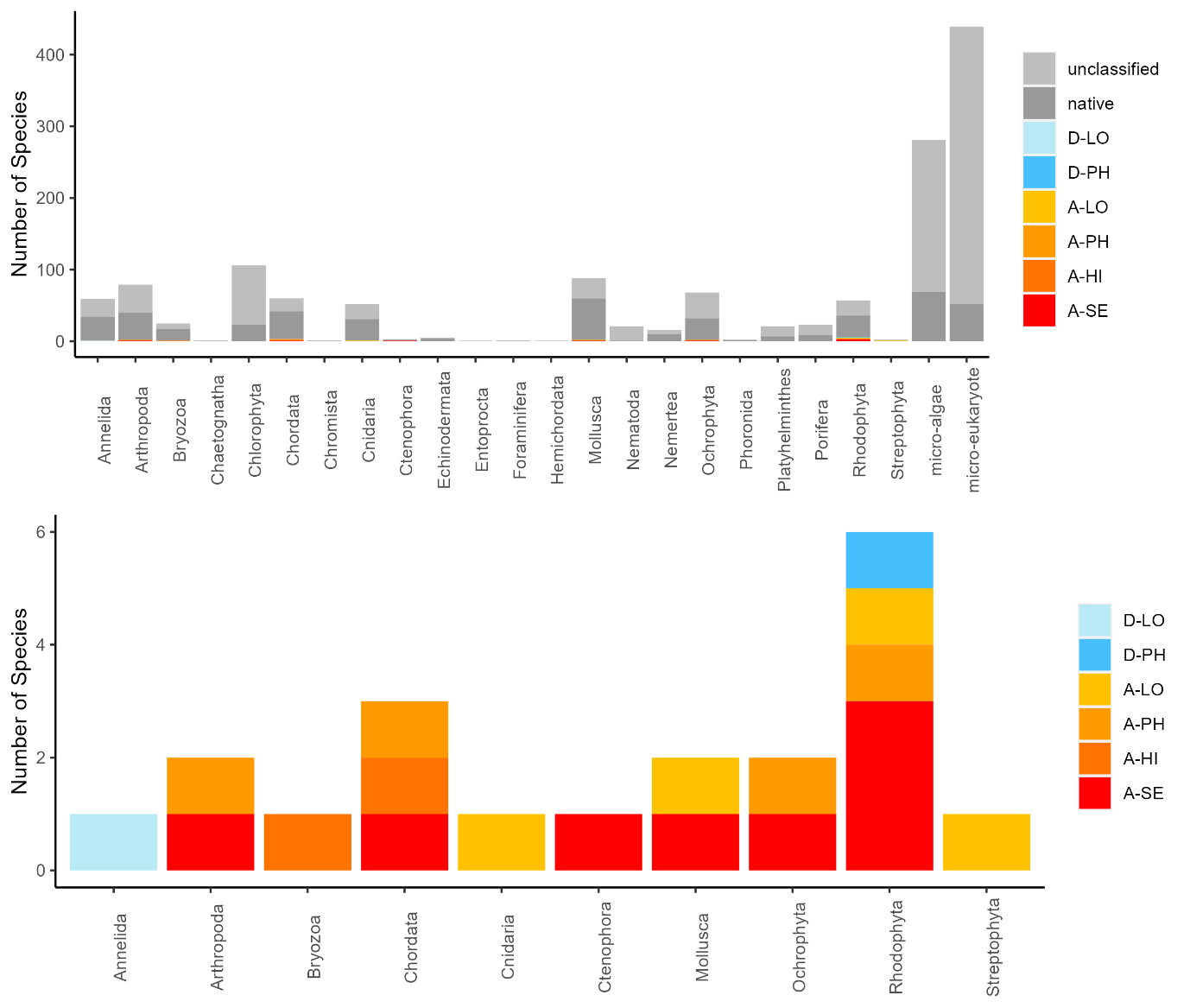
The conventional analyses recorded 37 species from the zooplankton samples. DNA-metabarcoding recorded 148 species belonging to the same phyla investigated by conventional methods and an additional 192 species belonging primarily to macro-algal, micro-eukaryote and micro-algal phyla that were not investigated in conventional surveys ( Figure 11 ). Among these were 14 marine alien species, of which two species ( Penilia avirostris and Caprella mutica ) were recorded by conventional methods while all species were recorded by DNA metabarcoding ( Figure 12 ). However, only a limited group of organisms was investigated by conventional methods as compared to DNA-based methods (Arthropoda, Chordata, Mollusca, Cnidaria, Foraminifera, Annelida, Chaetognatha). The eDNA method consistently detected more species in each of these groups than observed in conventional surveys. Half of the alien species detected using DNA metabarcoding belonged to Ochrophyta and Rhodophyta, two phyla that were not targeted in conventional analyses of the samples.
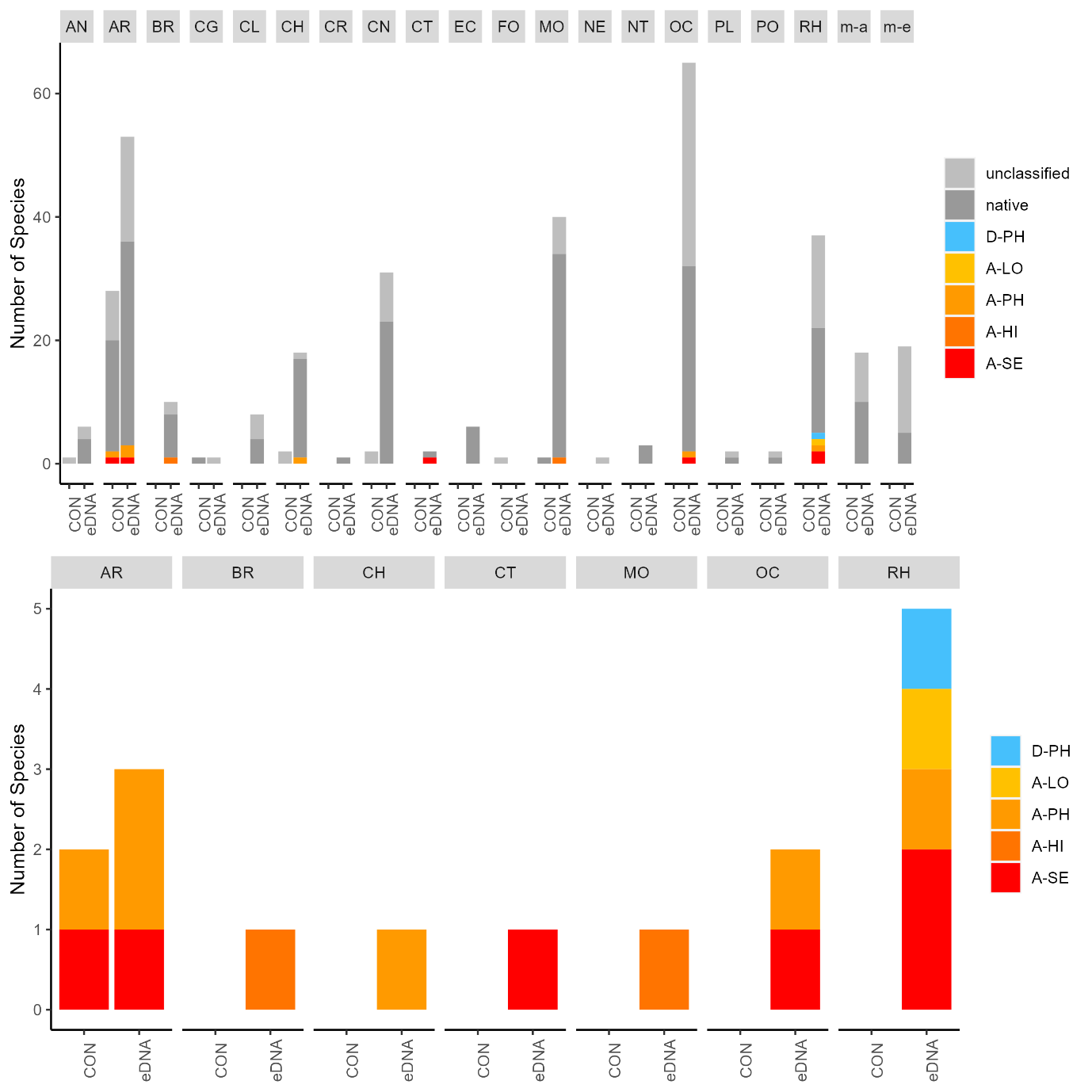
3.2.2 - Infauna in sediments
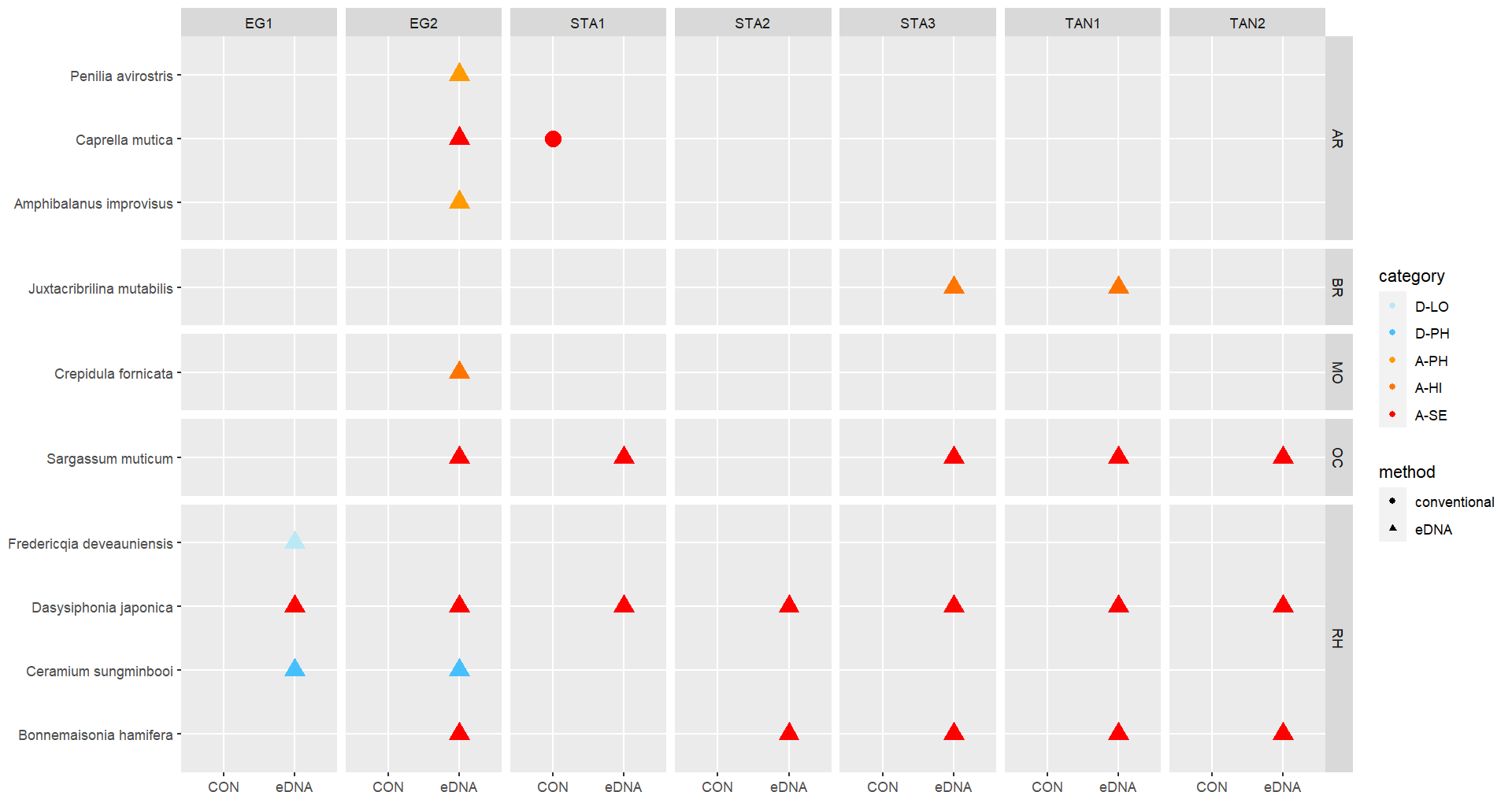
Conventional analyses of sediment samples recorded 99 species in total. DNA-metabarcoding detected 117 species belonging to the same phyla investigated by conventional analyses, and an additional 227 species belonging primarily micro-eukaryote and micro-algal phyla that were not investigated in conventional surveys ( Figure 13 ). Ten alien species were recovered from these samples, all of which were detected by DNA metabarcoding. Only one of these alien species ( Caprella mutica ) was recorded by conventional methods ( Figure 14 ). Except for Fredericqia deveauniensis , all alien species detected in sediments were also detected using other sampling methods (see Figure 25).
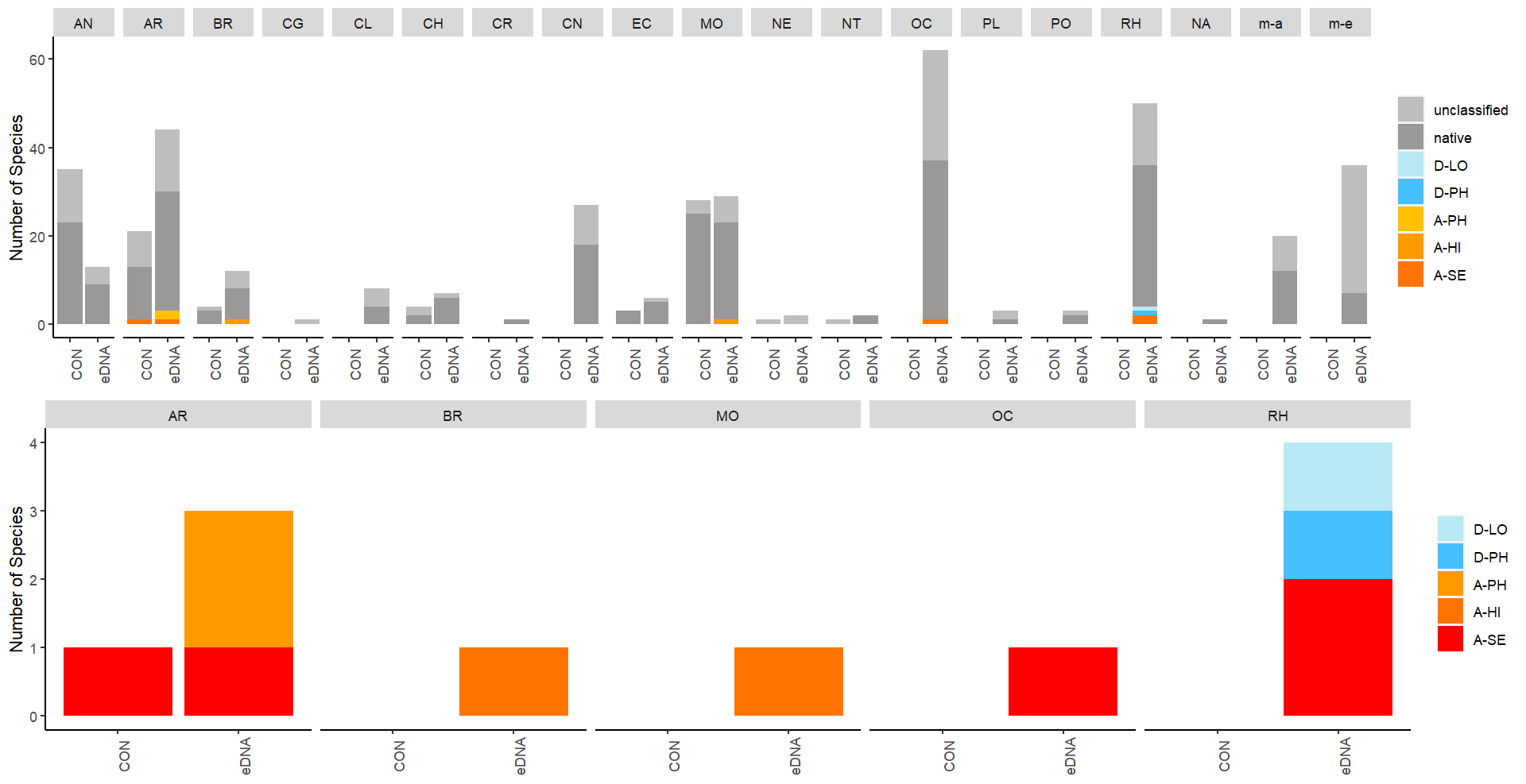
3.2.3 - Settlement panels
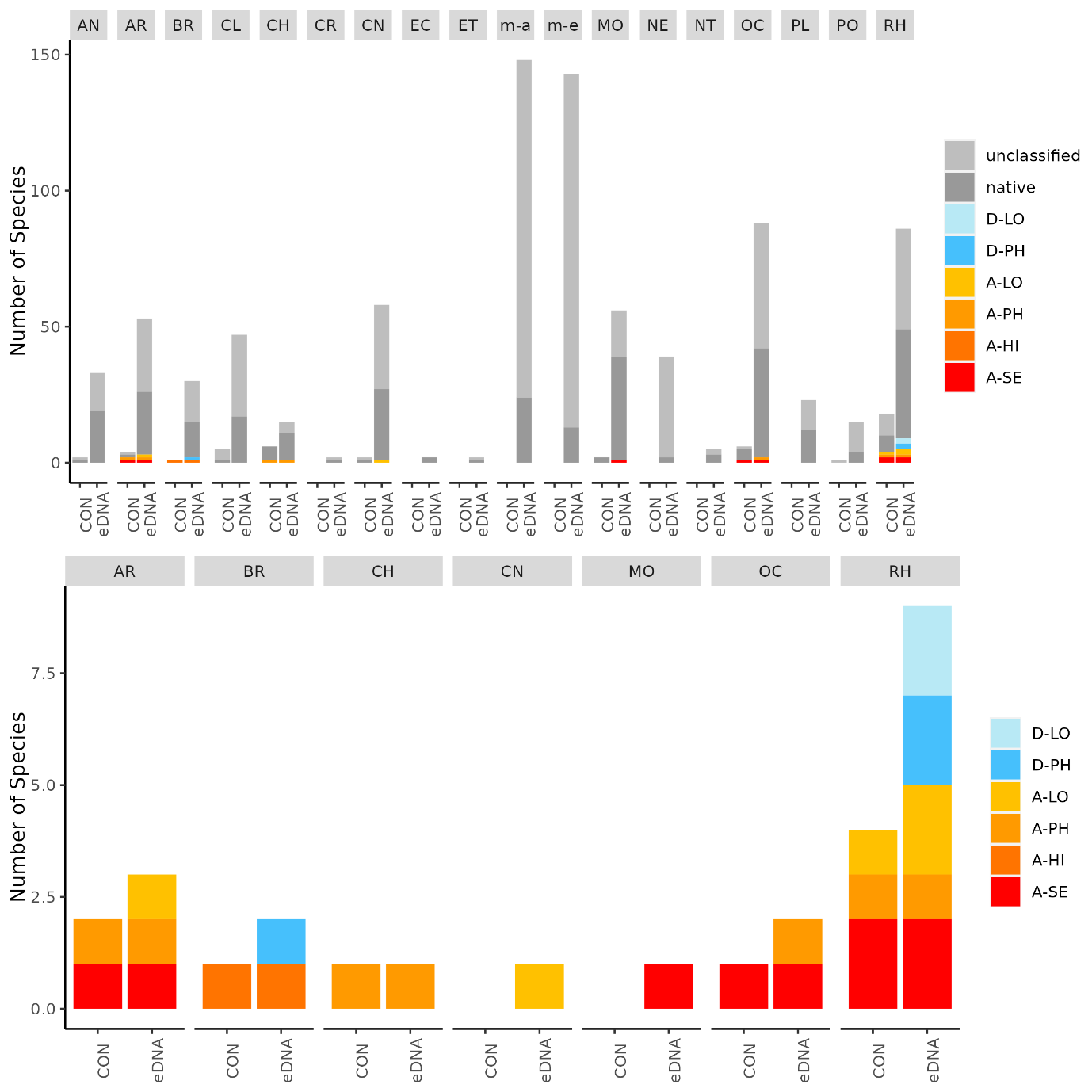
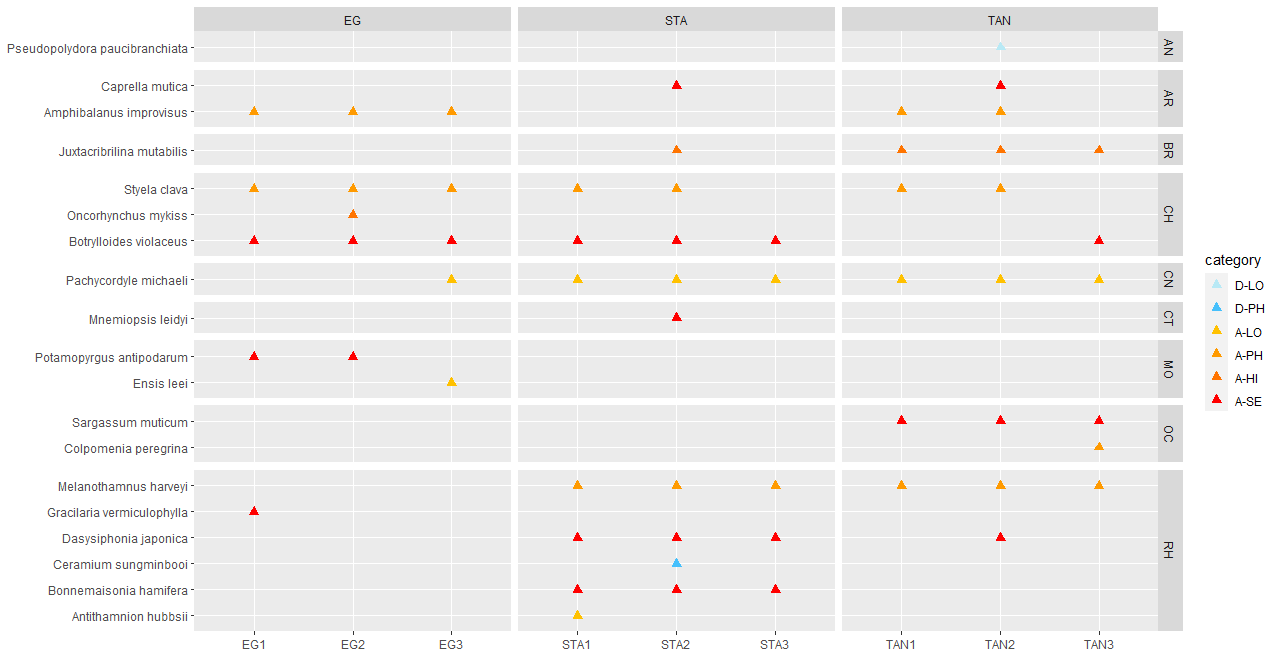
Conventional analyses of settlement panel specimens recorded in total 47 species. DNA-metabarcoding of the same panels recorded 478 species belonging to the same phyla recorded by conventional means, in addition to 503 species belonging primarily to micro-eukaryote and micro-algal phyla not investigated with conventional analyses ( Figure 16 ). The settlement panels recorded 20 marine alien species in total, of which 10 species were recorded by conventional methods and 18 species were recorded by DNA metabarcoding ( Figure 17 ).
Some of the settlement panels were completely covered by native ascidians when retrieved while some were rather bare ( Figure 15 ). The panels had a cover of feces from ascidians and a layer of organic material that were almost impossible to identify in the lab.
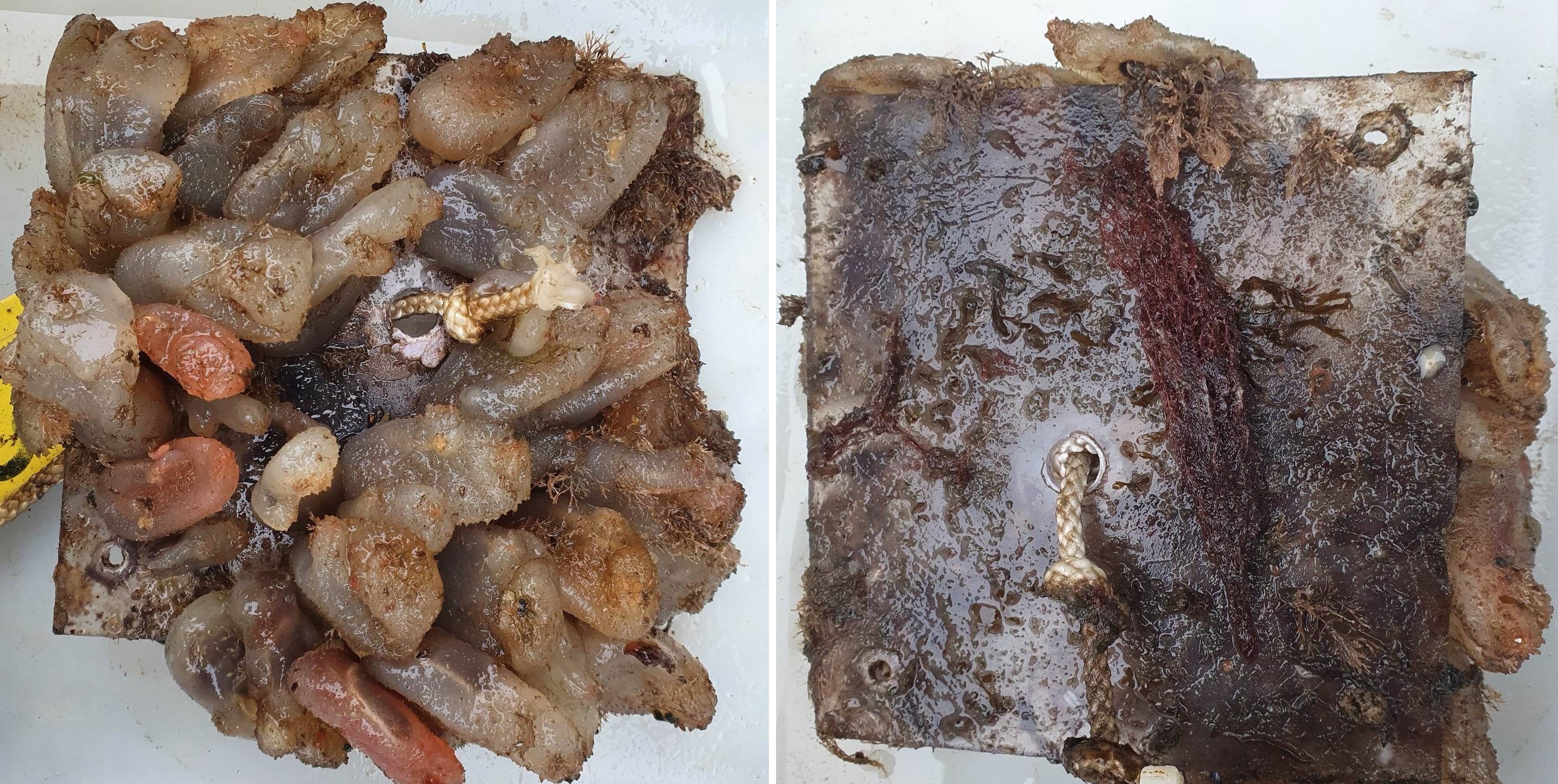
In the Stavanger area, only one of three settlement rigs was intact when collected, while in the other areas all rigs were retrieved. Although this would be expected to create some bias in the data, similar total numbers of species (EG: 597, STA: 474, TAN: 754) and alien species (EG: 19, STA: 16, TAN:18) were recovered from all three areas. However, there is a pronounced reduction in the number of alien species detected in Stavanger by conventional analyses (EG: 9, STA:4, TAN: 7) ( Figure 16 ). Although there was variability in the detection of marine alien species from panels from different depths, there was not a clear signal of only specific species being found at specific depths ( Figure 17 ).
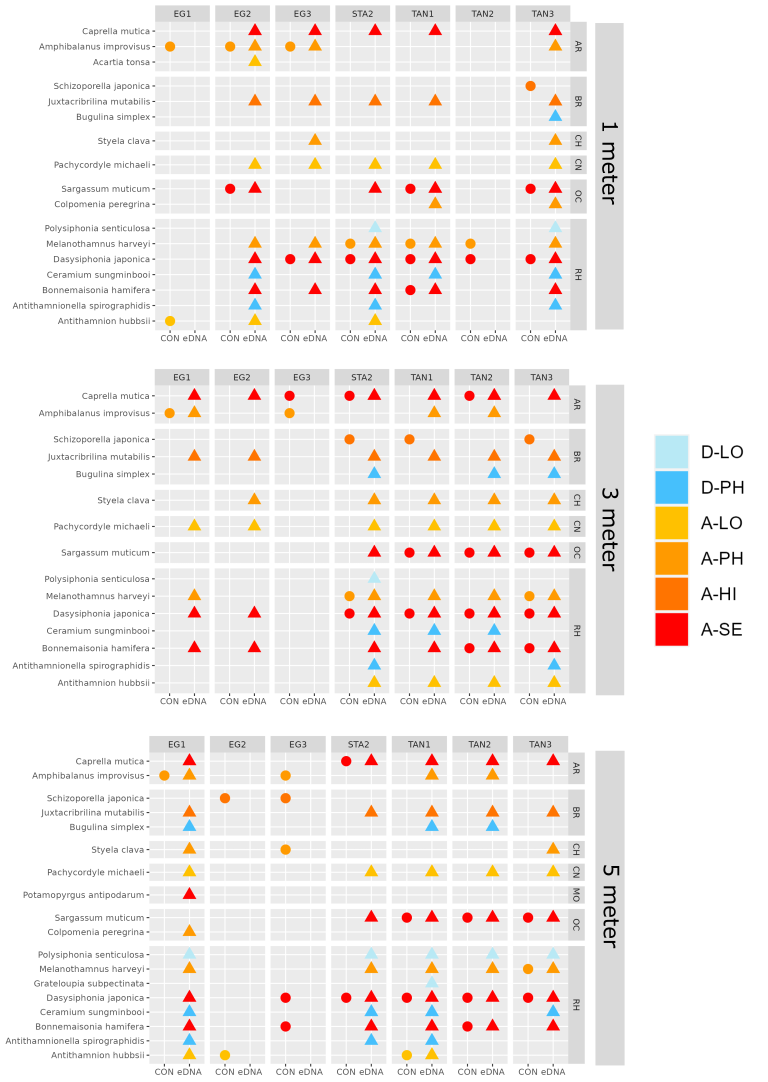
3.2.4 - e-DNA water samples
The eDNA water samples recorded 1705 species in total of which 662 belonged to phyla investigated by conventional methods. The remaining taxa were primarily micro-eukaryotes, micro-algae, and microbes. Among the taxa detected in water samples were 19 marine alien species ( Figure 18 , Figure 19 ). When a species was detected in an area, it was often detected in all 3 of the water samples taken in that area (12 of 30 cases; Figure 19 ).
3.2.5 - Crab pots and minnow traps
Fishing with crab pots and minnow traps yielded no alien species in either of the ports. The traditional crab pots had very low catches, while the minnow traps fished well. European green crab ( Carcinus maenas ), black gobies ( Gobius niger ), shrimps ( Palamon spp.) were the most frequent organisms in the traps, with some other fishes, snails and crabs present as well.
3.2.6 - Dredging and video transects
A total of 66 species were recorded by dredging, of which seven were alien species ( Table 2 ). This was the only method that physically collected the red algae Agarophyton vermiculophyllum in Egersund (EG 3) and the invasive ascidian Didemnum vexillum in Stavanger port (STA 1). Analysis of the video-transects did not yield any confident identification of alien species.
Table 2 . Alien species collected by dredging in the ports of Stavanger (STA), Tananger (TAN) and Egersund (EG). Numbers given are number of sites the species was recorded on.
| Latin name | Norwegian name | STA/rec sites | TAN/rec sites | EG/rec sites |
| Agarophyton vermiculophylla | 0 | 0 | 1 | |
| Dasysiphonia japonica | Japansk sjølyng | 2 | 0 | 0 |
| Melanothamnus harveyi | 1 | 1 | 0 | |
| Sargassum muticum | Japansk drivtang | 3 | 1 | 0 |
| Bonnemaisonia hamifera | Rødlo | 3 | 1 | 0 |
| Didemnum vexillum | Japansk sjøpung, havnespy | 1 | 0 | 0 |
| Schizoporella japonica | 0 | 1 | 1 |
3.2.7 - Rapid coastal survey (RCS)
Studies of the fauna on floating docks and ropes in marinas recorded 112 species of which 14 were alien species ( Table 3 ). This method was the only conventional method that recorded the alien bryozoan Tricellaria inopinata and red seaweed Grateloupia turuturu, both species found exclusively at the Stavanger sites.
Table 3 . Alien species collected in marina investigations/ Rapid Coastal Survey (RCS) in the ports of Stavanger (STA), Tananger (TAN) and Egersund (EG). Numbers given are number of sites the species was recorded on.
| Latin name | Norwegian name | STA/rec sites | TAN/recsites | EG/recsites |
| Antithamnion hubbsii/nipponicum | 2 | 1 | 0 | |
| Bonnemaisonia hamifera | Rødlo | 3 | 2 | 0 |
| Ceramium cf. sungminbooi | 3 | 0 | 0 | |
| Codium fragile | Pollpryd | 4 | 0 | 0 |
| Dasysiphonia japonica | Japansk sjølyng | 2 | 0 | 1 |
| Grateloupia turuturu | Djevletunge | 4 | 0 | 0 |
| Melanothamnus harveyi | 3 | 2 | 0 | |
| Sargassum muticum | Japansk drivtang | 3 | 3 | 1 |
| Amphibalanus improvisus | Brakkvannsrur | 0 | 0 | 3 |
| Caprella muticum | Japansk spøkelseskreps | 3 | 2 | 0 |
| Mnemiopsis leidyi | Amerikansk lobemanet | 4 | 0 | 0 |
| Styela clava | Lærsekkdyr | 2 | 3 | 1 |
| Schizoporella japonica | 2 | 3 | 1 | |
| Tricellaria inopinata | 2 | 0 | 0 |
3.2.8 - Beach survey
Snorkeling, beach survey, prawn nets and collection of fauna associated with fucoid plants reviled 74 species of which nine species were alien to Norway ( Table 4 ). In Stavanger port there were no actual beaches at the sites studied and only short snorkelling transects were performed along artificial shore and no alien species were detected.
Table 4 . Alien species collected in beach survey/snorkeling in the ports of Stavanger (STA), Tananger (TAN) and Egersund (EG). Numbers given are number of sites the species was recorded on.
| Latin name | Norwegian name | STA/rec sites | TAN/recsites | EG/rec sites |
| Dasysiphonia japonica | Japansk sjølyng | 0 | 1 | 0 |
| Melanothamnus harveyi | 0 | 2 | 0 | |
| Sargassum muticum | Japansk drivtang | 0 | 2 | 0 |
| Bonnemaisonia hamifera | Rødlo | 0 | 1 | 1 |
| Crepidula fornicata | Tøffelsnegl | 0 | 1 | 0 |
| Mnemiopsis leidyi | Amerikansk lobemanet | 0 | 0 | 3 |
| Styela clava | Lærsekkdyr | 0 | 1 | 0 |
| Amphibalanus improvisus | Brakkvannsrur | 0 | 0 | 1 |
| Magellana gigas | Stillehavsøsters | 0 | 2 | 1 |
3.3 - Marine alien species collected in this study
A total of 26 marine alien species and seven risk-assessed doorknocker species (33 in total) were detected in this study. Of the seven doorknocker species two of them have been recorded once in Norway before, while five have never been recorded. Some species were only found by conventional methods (7), and some were exclusively detected by DNA analysis (14) while the remaining species (12) were detected by both methods ( Table 5 ). The DNA methods detected alien species at more localities and was more consistent in recording the same species across all stations in an area (see section 3.4 below).
From the conventional analyses, a total of 307 marine species were recorded in this study, of which 19 were marine alien species or risk-assessed doorknocker species. From the DNA-based analyses, a total of 2274 species were recorded, of which 1014 belonged to phyla investigated using conventional methods. The remaining taxa belonged primarily to micro-eukaryote and micro-algal groups that were not investigated by taxonomic experts using conventional methods. The taxa detected included 26 alien marine species and doorknocker species. 653 species could be confirmed as native to Norway, and 1594 were considered unclassified. Among the unclassified species were 634 taxa that could not be identified to the species level and 686 taxa that did not belong to phyla investigated using conventional methods (i.e., primarily micro-eukaryote, micro-algal, and microbial taxa that also have not been systematically risk assessed as alien species). The remaining 274 unclassified species represent a combination of species that may be i) species native to Norway that have been previously overlooked, ii) taxonomic irregularities or synonyms of native Norwegian species that are not registered in the Norwegian Species Nomenclature Database, iii) incorrectly identified species native to Norway, and iv) new introduced alien species to Norway’s coastal waters (Appendix 3).
Table 5 . Marine alien species recorded by conventional and DNA methods in this study. Risk category from the Norwegian Biodiversity centre: SE- very high risk, HI-high risk, PH- potential high risk, LO low risk. Species marked * is only found by conventional methods, species marked ** are only found with DNA based analysis. Species marked by † are detected only by DNA based analysis and were initially missed in automatic screening of species lists as they were identified under a taxonomic synonym that did not appear in the Norwegian Species Nomenclature Database and required manual curation by taxonomic experts to confirm them as alien species.
| Species | Author name | Origin | Year of arrival | Risk category | |
| Acartia tonsa** | Dana, 1849 | Crustacea | North Pacific | 2012 | LO |
| Agarophyton vermiculophyllum (Gracilaria vermiculophylla) | (Ohmi) Gurgel, J.N. Norris & Fredericq, 2018 | Rhodophyta | North Pacific | 2012 | SE |
| Antithamnion hubbsii (nipponicum) | E.Y. Dawson, 1962 | Rhodophyta | North Pacific | 2007 | LO |
| Antithamnionella spirographidis† | (Schiffner) E.M. Wollaston 1968 | Rhodophyta | Uncertain | Door knocker | PH |
| Amphibalanus improvisus | (Darwin, 1854) | Crustacea | West Atlantic | 1900 | PH |
| Bonnemaisonia hamifera | Hariot | Rhodophyta | North Pacific | 1902 | SE |
| Botrylloides violaceus** | Oka, 1927 | Ascidiacea | North Pacific | 2007 | SE |
| Bugulina simplex** | Hincks,1886 | Bryozoa | Mediterranean | Door knocker | PH |
| Caprella mutica | Schurin, 1935 | Crustacea | North Pacific | 1999 | SE |
| Ceramium sungminbooi | J. R. Hughey & G. H. Boo | Rhodophyta | North Pacific | Door knocker (1996) | PH |
| Codium fragile * | (Suringar) Hariot | Chlorophyta | North Pacific | 1932 | SE |
| Crepidula fornicata | (L. 1758) | Mollusca | West Atlantic | 1958 | HI |
| Colpomenia peregrina** | Sauvageau | Phaeophyta | North Pacific | 1933 | PH |
| Dasysiphonia japonica | (Yendo) H.-S. Kim | Rhodophyta | North Pacific | 1996 | SE |
| Didemnum vexillum* | Kott, 2002 | Ascidiacea | North Pacific | 2020 | SE |
| Ensis leei ** (Ensis directus) | Huber, 2015 | Mollusca | West Atlantic | 1989 | LO |
| Fredericqia deveauniensis** | Maggs, L. LE Gall, Mineur, Provan & G.W. Saunders, 2013 | Rhodophyta | Uncertain | Door knocker (2016) | LO |
| Grateloupia subpectinata† | Holmes 1912 | Rhodophyta | Uncertain | Door knocker | LO |
| Grateloupia turuturu* | Yamada, 1941 | Rhodophyta | North Pacific | 2019 | HI |
| Juxtacribrilina mutabilis ** | Ito, M.; Onishi, T.; Dick, M. H. (2015). | Bryozoa | North Pacific | 2008 | HI |
| Magellana gigas (Crassostera gigas) * | (Thunberg 1973) | Mollusca | North Pacific | 2007 | SE |
| Melanothamnus harveyi | (Bailey) Díaz-Tapia & Maggs | Rhodophyta | West Atlantic | 1983 | PH |
| Mnemiopsis leidyi* | A. Agassiz, 1865 | Ctenophora | West Atlantic | 2005 | SE |
| Oncorhyncus mykiss ** | Walbaum, 1792 | Chordata | West Atlantic | 1902 | HI |
| Pachycordyle michaeli ** | (Berrill, 1948) | Cnidaria | Uncertain | 2018 | LO |
| Penilia avirostris | Dana, 1849 | Arthropoda | Cosmo | 2002 | PH |
| Polysiphonia senticulosa** | Harvey, 1862 | Rhodophyta | West Atlantic | Door knocker | LO |
| Potamopyrgus antipodarum ** | Gray, J. E. (1843) | Mollusca | New Zealand | 1954 | SE |
| Pseudopolydora paucibranchiata ** | Okuda, 1937 | Polychaeta | North Pacific | Door knocker | NA |
| Sargassum muticum | (Yendo) Fensholt | Phaeophyceae | North Pacific | 1988 | SE |
| Schizoporella japonica * | Ortmann, 1890 | Bryozoa | North Pacific | 2014 | HI |
| Styela clava | Herdman, 1881 | Tunicata | North Pacific | 1990 | PH |
| Tricellaria inopinata* | d'Hondt & Occhipinti Ambrogi, 1985 | Bryozoa | Pacific | 2014 | SE |
3.3.1 - The seaweeds
The green alga Codium fragile , the red alga Bonnemaisonia hamifera and brown alga Colpomenia peregrina has been in Norway for a long time and are widely distributed. The brown alga Sargassum muticum , the red algae Melanothamnus harveyi and Dasysiphonia japonica arrived in the eighties and nineties and are well established along the coast. In this study these species were recorded by all methods except from traps, sediment, and zooplankton samples. Another species of Melanothamnus was detected from all DNA analysis; M. japonicus . M. harveyi and M. japonicus are confirmed separate species by Savoi & Saunders (2015), however, the origin of these species is still unclear (van der Loos & Bafort, et al. 2023).
The red algae Antithamnion hubbsii / nipponicum has also some taxonomic unclarities while some recognize them as two species and some as the same species (van der Loos & Bafort, et al. 2023). Antithamnion hubbsii was first reported from Norway as A. nipponicum in 2007 (Rueness et al. 2007). The present study registered A. hubbsii both by conventional methods and DNA-analysis in all areas.
A similar small red alga Antithamnionella spirographidis was detected by DNA analysis from settlement panels in all three ports. The species has an unclear origin but originates probably from Japan and was introduced to the Mediterranean before 1882 and was described from Italy in 1914. The species is now widespread in the North Sea but is a doorknocker to the Norwegian coast.
The red alga Grateloupia turuturu (Devils tongue) was recorded for the first time in Norway near Larvik in 2019. In 2021 the species was also found at two sites in Rogaland County (Mosterøy and Dusavika). In the present study the species was found to be abundant at all marinas studied in Stavanger port, suggesting a rapid dispersal ( Figure 20 ).
Another alga in the genus Grateloupia ; G. subpectinata , was detected in the DNA analysis from one settlement panel in the port of Tananger. The species is originally described from Japan and has been introduced to the Mediterranean Sea and spread to western European coasts. The species is a doorknocker to Norway (Husa et. Al. 2023).
The red alga Ceramium sungminbooi was recorded by DNA-analysis of herbarium specimen identified as Ceramium cimbricum collected in Norway in 1996. The species is recently described from Korea in 2016 ( Hughey, J.R. & Boo, G.H. 2016). The species is very similar to
C. cimbricum and misidentification is plausible. In the present study we detected C. sungminbooi by DNA analysis on settlement panels, and zooplankton samples from all ports, from sediment samples in Egersund and from water samples in Stavanger. The examination of Ceramium species in the lab resulted in several specimens that could not be identified to species level with confidence. None of the DNA analysis gave signal of C. cimbricum , which indicates that C. sungminbooi is a widespread species in this area.

In the present study the red alga Agarophyton vermiculophyllum (Gracilaria vermiculophylla) was found for the first time on the west coast at one station in Egersund (EG3). eDNA from water samples also indicated that the species is present at another site in Egersund (EG1) ( Figure 20 ). The species was first recorded from Tønsberg on the east coast in 2012 and has spread rapidly along the coast from the Swedish border to Kristiansund (Husa et al. 2023).

The red alga Polysiphonia senticulosa has not been observed in Norway before but was detected with DNA analysis of settlement panels in all the areas. P. senticulosa is native to the western Atlantic coast. The species was introduced to the Netherlands in 1993 and has spread to Belgium and France. P. senticulosa is very similar to native P. stricta and may have been overlooked in Norway (Husa 2023). There were several Polysiphonia sp. in the material collected, which should be studied further for possible species identification. Fredericqia deveauniensis is a small red alga, with one erect and one crust stage. The species is of an uncertain origin but is present in Ireland and along the eastern Canadien shores. The crust stage was found in Norway only once in 2016 (Bringloe et al. 2016). The species is easily confused with native red alga. In this study the species was detected by DNA analysis of sediment samples in the port of Egersund.
3.3.2 - The fauna
The ascidian Botrylloides violaceus was first registered in Norway in 2007 (Egersund area). The species is very difficult to distinguish from native B. leachii in laboratory unless it is recognized by red larvae in the spring ( Figure 22 ). B. violaceus originates from Japan and was first observed in Norway from several sites in Egersund. In this study we could not confirm the species by morphology, but DNA-analysis detected B. violaceus from water samples in all ports.
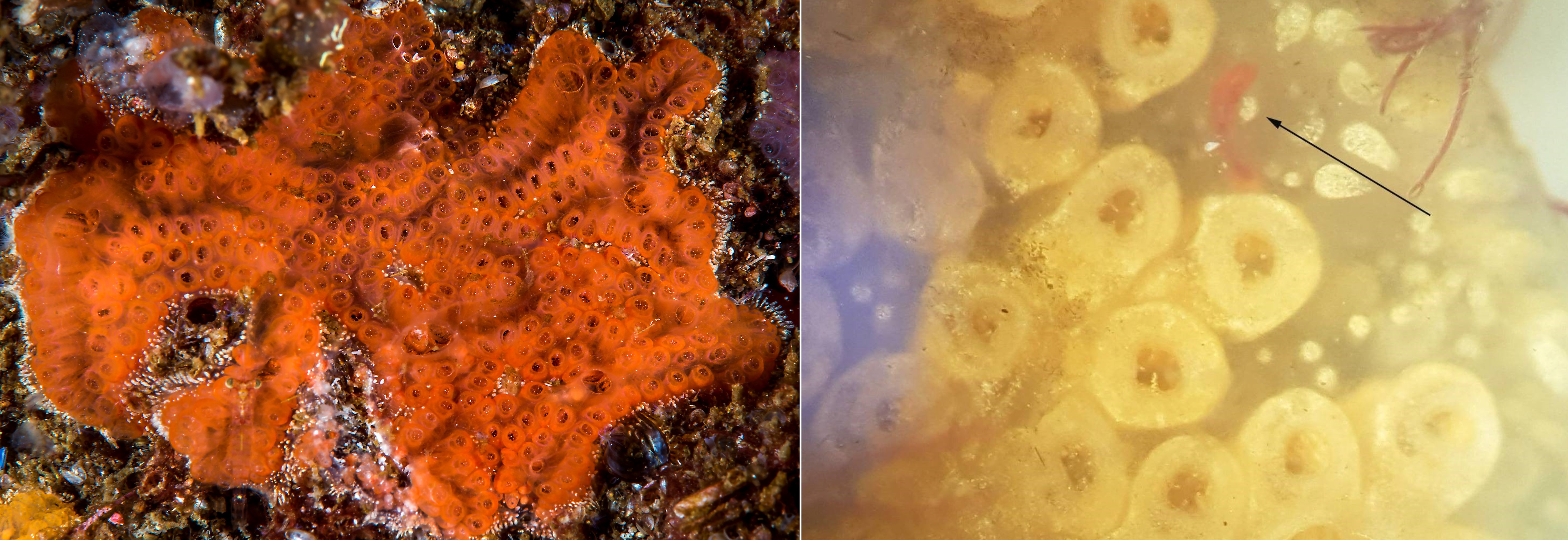
The invasive ascidian Didemnum vexillum was only recorded at one site (STA 1) in Stavanger port, where a small colony was found on loose laying Fucus serratus . The DNA analysis did not detect any signal from D. vexillum , even if the species is known to be present near STA 1 and STA 3 in Stavanger port and at EG 3 in Egersund.
Pacific oyster Magellana gigas were found at one settlement panel in Egersund port and at two sites in Tananger port and one site in Egersund during the beach surveys.
The snail Potamopyrgus antipodarum was only detected by DNA samples from the settlement panels in Egersund (EG1) which is the site closest to the river and most freshwater influenced. P. antiporadum thrives in freshwater and brackish water and has been found in disjunct rivers and lakes since 1954 but has never been recorded from Rogaland County.
The slipper limpet Crepidula fornicata was registered by snorkelling in Tananger and detected by DNA analysis both in the sediment samples and the zooplankton samples from Egersund.
C. fornicata has earlier been registered with sporadic specimens in Rogaland.
The ctenophore Mnemiopsis leidy was observed during the RCS at all stations in Stavanger port and from the genetic analysis of zooplankton and waters samples from the same area. M. leidy has been present in Norwegian waters since 2005 and has formed regular blooms in August to December along the coast up to Trondheimsfjorden (Falkenhaug et al. 2023).
The American razor clam Ensis leei was detected by e- DNA from waters samples from Egersund port, but not found in the beach surveys. The species was first recorded in Norway in 1989 and has spread along the Skagerrak coast.
Pachycordyle michaeli is a small hydromedusa which was first discovered in Norway in 2018 and with just one more record in 2019 (Falkenhaug et al. 2023). In this study the species was only recorded from e-DNA in water samples in Tananger.
Acartia tonsa is a small copepod that thrives in freshwater and brackish water. A. tonsa was only detected by DNA analysis of settlement panels in Egersund. The species was registered in Norway for the first time in 2012. The distribution of the species in Norway is poorly known, but it has been observed and detected by DNA analysis along the Skagerrak coast (Moseid et al. 2021; Falkenhaug et al. 2023).
Penilia avirostris is a small sub-tropic crustacean (Cladocera) present in most of the world's coastal areas. P. avirostris was first recorded from the Norwegian coast in 2002 and is now commonly found in plankton samples from the Skagerrak coast (Falkenhaug et al. 2023). In this study the species was found in the plankton samples and in the DNA analysis of zooplankton and sediment samples from Stavanger and Egersund port.
The bryozoan Bugulina simplex ( Figure 23 ) was only detected by DNA analysis of the settlement plates, but signals were present in all the ports examined. B. simplex is introduced from the Mediterranean Sea to countries in western Europe (Belgium, Netherland, Shetland and the British Isles). Specimens suspected to be B. simplex were found in 2018 in Stavanger (Benjaminsen 2022).
Juxtacribrilina mutabilis ( Figure 23 ) is a small pink crust bryozoan originating from the North Pacific and has been introduced to USA, Sweden and Norway. The first findings in Norway were in 2008 and the species was found on nine sites in an investigation in 2018 (Benjaminsen 2022). The species is small and probably overlooked in Norway (Husa et al. 2023). In this study J. mutabilis was detected by DNA analysis of zooplankton and sediment samples in Tananger and Stavanger port.
Schizoporella japonica ( Figure 23 ) was identified from the settlement panels and from marinas in all three ports but was not detected by DNA analysis. However, Schizoporella dunkeri gave signal in the DNA analysis, and it is impossible to separate these two species based on information given in the gene banks. S. japonica was first discovered in Norway in 2014 and has spread quickly along the coast (Husa et al. 2023).
Tricellaria inopinata was recorded from two marinas in Stavanger port in this study. The species was first recorded i Florø in 2014 (Porter et al. 2015).

Pseudopolydora paucibranchiata is a polychaete worm that previously have been reported as common along Norwegian shores. A revision of the genus shows that this species is an alien species from Japan and that the Norwegian recordings are misidentified and should probably belong to P. nordica (Radashevsky 2001). The species is not evaluated for the Norwegian alien species list 2023. This study detected P. paucibranchiata in water samples from Tananger port.
Rainbow trout, Oncorhynchus mykiss , was detected in eDNA samples from Egersund. This anadrome salmonid fish of west Atlantic origin was introduced and released in Norway in the early 1900’s, though today there are not many rivers or lakes with established reproducing populations. The species is used in aquaculture in the coastal zone, from which there are escapees (Forsgren et al. 2023a).
3.3.3 - Patogenes in oysters
Several mud blisters were found in Pacific oyster in Tananger port, but we were not able to detect any worms in the blisters. Additional sampling has been performed to do DNA analysis of the mud in the blisters ( Figure 24 ).

3.4 - Potential introductions detected by DNA-methods.
Among those 1640 taxa identified to the species level from the metabarcoding data, 653 represented native species and 26 represented known alien or doorknocker species from the Norwegian Invasive Species List. The remaining 961 species are either registered as “Not Found in Norway” in the Norwegian Species Nomenclature Database or are not included in the database at all. Many of these species (686) are micro-algal, micro-eukaryote, or microbial taxa that were not investigated using conventional methods and are typically not assessed in alien species lists.
These unclassified species include a mixture of i) species native to Norway that have been previously overlooked, ii) taxonomic irregularities or synonyms of native Norwegian species that are not registered in the Norwegian Species Nomenclature Database, iii) incorrectly identified species native to Norway, and iv) new introduced alien species to Norway’s coastal waters. We have curated this list to identify species possibly belonging to groups i, ii, and iv (Appendix 3) but have not systematically screened the data for species that may have been misidentified using the genetic methods.
Our screening of the unclassified species identified two synonyms representing an additional 2 doorknocker species from the Norwegian Invasive Species list: Grateloupia subpectinata and Antithamnionella spirographidis . Sixty-four of the species were identified as synonyms or mis-spelled native species, and an additional twenty-one species were classed as potentially overlooked native taxa, as they are known to occur within Scandinavian waters. One hundred and seventy species were identified as potential introductions that warrant further investigation.
With further development of custom identification databases and the use of probabilistic classifiers for DNA-based species identification, it is possible to assess marker performance on a genus or family level to provide improved confidence measures on species identifications and thereby distinguish between likely misidentifications and good candidates for species that represent new introductions to Norwegian waters.
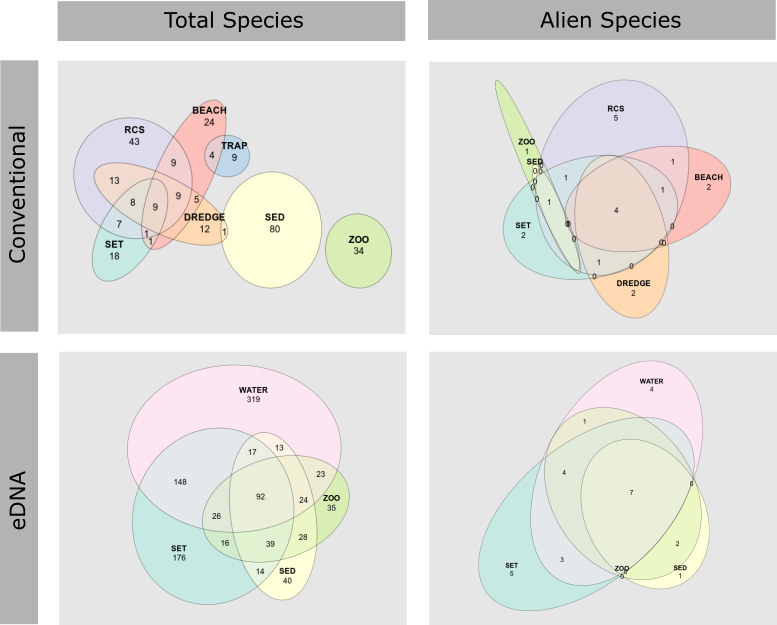
3.5 - Comparison of field methods for assessing marine alien species
The highest number of species in general were found in the sediment and RCS by the conventional methods, and in the water and settlement plates by the genetic methods ( Figure 25 , Figure 26 ). The overlap in species records among collection methods were far higher using genetic than conventional species identification ( Figure 26 ). This is due to multiple markers targeting different taxonomic groups being analyzed for most sample types, whereas the taxonomical experts only analyzed selected target groups for each sample type.
The highest number of alien species were found in RCS, settlement plates, and beach surveys by the conventional methods, and in the settlement plates and water samples by genetic methods Figure 25, Figure 26).
3.6 - Comparison of genetic markers for assessing marine alien species
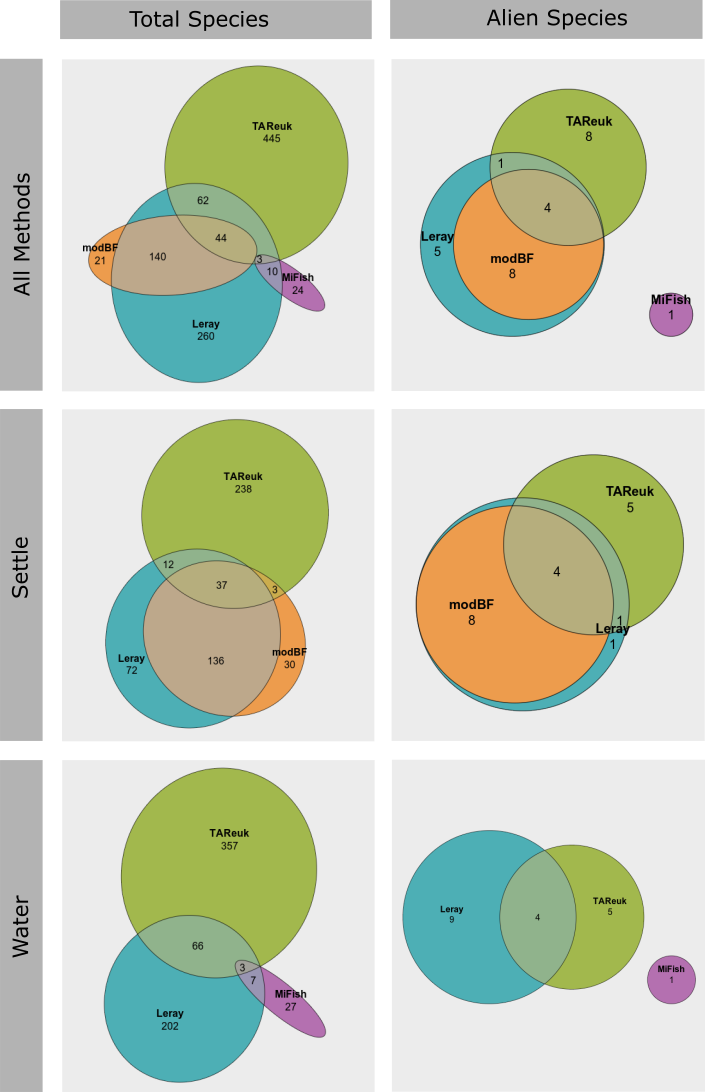
We only selected one single genetic marker per taxonomic group in this study, except for two different markers targeting invertebrates (Leray and modBF) for the settlement plate samples. We see that modBF do find some unique species in addition to the species also found by the Leray marker and vice versa ( Figure 27 ). But when looking at alien species only, there are only a few unique species. We do however expect that multiple markers targeting the same taxonomic group will significantly increase the number of species registered.
For the zooplankton and sediment samples, only a single genetic marker was analysed, and we cannot make any comparisons for these sample types. But for the settlement plates and water samples, we see that the different genetic markers targeting different taxonomic groups provide unique species as expected ( Figure 27 ). The main markers selected for this study represent algae (TAReuk), invertebrates (Leray and modBF) and fish/vertebrates (MiFish), and this provides information on very different taxonomic groups.
4 - Discussion
4.1 - General findings
A combination of different field collection methods covering a wide range of habitats and implementing DNA metabarcoding in addition to conventional analyses returned a high number of marine alien species in the investigated harbours. A total of 33 alien species were detected and DNA analysis indicates that several new door knocker species are already present in Norwegian waters.
From the conventional analyses, a total of 307 marine species were recorded in this study, of which 19 were marine alien species or risk-assessed doorknocker species. From the DNA-based analyses, a total of 2268 species were recorded, of which 1014 belonged to phyla also investigated using conventional methods. The DNA-based analyses detected 26 alien marine species and doorknocker species. Importantly, the DNA-based analyses were more consistent where specific alien species were found more often within and between both samples and sites, likely resulting in a much higher probability of detection.
A recent Danish study, known as the MONIS-4 report (Andersen et al. 2022), recorded 34 alien species across 16 harbours, where 26 species were found by conventional methods and 13 species were found by DNA based methods. However, the Danish approach did not include DNA-metabarcoding, but rather targeted 18 specific alien species using species-specific genetic markers. Hence, any other alien species than these 18 could not be detected with the genetic analyses. A screening of marine litter and settlement plates in the Netherlands using DNA metabarcoding recorded 46 non-indigenous algae and animals, from samples during three consecutive years (Polling et al. 2023). The pilot study conducted in only 3 harbours in Norway therefore found a comparably high number of alien species.
4.1.1 - Selection of field collection methods for detection of marine alien species
As part of this study, several different collection methods were utilized across all sample sites to compare the efficacy and potential overlap and redundancy among them. Based on both the total number of species and the number of alien species detected by each method, it becomes clear that some collection methods detected relatively few species whereas other methods detected more species. In general, the studies of marinas (RCS) and water samples recorded the highest number of alien species independent of species identification method. However, the settlement plates and beach surveys also provided relatively high contributions to the alien species list using conventional methods.
Even though some methods detected few alien species, they still cover different habitats and different groups of species. Traps collect mobile fauna like pacific shore crabs and gobies, but there were none present in the study area. Collection of sediment may collect alien polychaetes and zooplankton samples covers this group better than any other conventional methods. Identifying species in sediments and zooplankton samples is time requiring and makes such investigations very costly. The use of dredge collected fewer species but still two species that were unique as the method were able to sample deeper than the other conventional methods. Video transects are better performed using an underwater drone to be able to confirm the extent of species of special interest, such as Didemnum vexillum and Agarophyton vermiculophyllum .
Importantly, most of the field collection methods tested provide little physical material at a single time point, with the settlement plates representing an exception. The long “sampling period” where these plates are collecting material from the sea increases the probability of detecting rare alien species. Hence, increasing the size of the physical material analysed either through increasing sample size per visit, or increasing the number of visits per year would likely increase the detection probability of alien species. One such possibility is to use “passive plankton samplers” (Tryggestad 2023) to increase both the sample size and sampling time to collect more representative samples for a specific area. Such passive samplers can be left in the sea for days or even weeks to collect large samples, increasing the probability of finding alien species. Whereas large physical samples can be a problem for conventional methods, as this will increase handing time and costs, the DNA-based methods are well suited for analysing larger samples.
The eDNA water samples provided the highest number of species in this study, but even these samples only represented a single time point and relatively few spatial points at each locality. Extending sampling in general to multiple times per year to include seasonal differences, and potentially also increasing the number of samples collected in each harbour would likely increase the number of alien species recorded. For the water samples for example, only surface samples were collected in this study, and including bottom samples would likely also increase the number of species recorded.
4.1.2 - DNA-based species identification compared to conventional analyses
The DNA-based methods identified both more species in general (2268 vs 307) as well as more alien species (26 vs 21) than the conventional methods in this study. The DNA-based methods also seemed to be more consistent, where the same alien species were detected in multiple samples within and among areas. DNA analysis additionally could confirm the presence of species that are difficult to distinguish from native species such as Antithamnion hubbsii , Ceramium sungminbooi and Botrylloides violaceus . By contrast, conventional analyses were unable to successfully detect and identify the invasive species Schizoporella japonica , while DNA based analyses seem to detect the species, but the marker used appears to incorrectly identify the species as Schizoporella dunkeri .
Whereas most conventional methods are based on morphology and taxonomical expertise, DNA-based methods are more general and less dependent on the knowledge of a globally diminishing number of experts. There is, however, a strong need for taxonomical expertise in interpreting and classifying the large amount of data created by DNA-based analyses. In this study, the genetic species identification relied on BLAST searches using the non-curated open data in GenBank, and this approach will often include false positive identifications. Curated reference databases such as MetaZooGene and probabilistic species identification approaches such as RDP classifier would provide fewer false positives.
The overlap in species records among collection methods were far higher using genetic than conventional species identification. However, the taxonomic experts only identified selected target groups for each sample type, whereas multiple markers targeting many taxonomic groups were analysed using genetic analyses. Hence the genetic analyses targeted the same taxonomic groups across different sample types, and it seems that most species can be detected even if some sample types were excluded. This is in accordance with previous studies where metabarcoding of planktonic larval stages was found to be an efficient approach for investigating benthic alien species (Couton et al. 2019).
The rapid development of eDNA and DNA metabarcoding has provided many genetic markers targeting different taxonomic groups across different environments. In this study we have selected only one single marker per taxonomic group, except for two different markers targeting invertebrates (Leray and modBF) for the settlement plate samples. Several other studies have shown that using multiple markers per taxonomic group will detect more species, and sometimes can find as much as 50% more species. Particularly for taxonomic groups with many alien species, including more than one marker could be valuable.
In this study, we only utilized general genetic markers and DNA metabarcoding. This method is excellent for analysing broad-scale biodiversity and screening for the presence of many species at the same time. However, DNA metabarcoding is quite time consuming in the genetic laboratory, it must be sent off to a sequencing centre for hi-throughput sequencing and it requires lots of bioinformatic analyses before a species list can be aggregated. For monitoring single alien species, such as sea vomit, species-specific genetic markers and qPCR analyses are commonly utilized to detect their presence (Fossøy et al. 2022). This approach is much faster in the genetic laboratory, and it returns an answer of presence or absence for a specific species without the need for any complex bioinformatic analyses. Hence, for monitoring the spread of single alien species, requiring many samples and quick results, qPCR would be a better option. In the Danish MONIS-4 study, qPCR was used to target 18 specific marine alien species (Andersen et al. 2022). However, this means that you only find the species you are looking for, and you could miss important alien species.
4.1.3 - Challenges of DNA-based Species Identification
DNA metabarcoding successfully detected many of the same species noted in conventional surveys, in addition to a broad array of additional species both within those organismal groups investigated using conventional methods, but also among other groups that were not investigated due to fiscal, logistical and/or time constraints (ex/Annelida, Nematoda, Cnidaria). However, large-scale database-reliant automated DNA-based species identification comes with an inherent error rate that must be taken into consideration when interpreting these detections. More specifically, the following issues can impact the quality and reliability of DNA-based species determinations:
4.1.3.1 - Marker resolution
-
To facilitate simultaneous identification of many species in a taxonomic group, specific marker regions are targeted. However, inter-species genetic variation can vary considerably between any two given species in a group. In some cases, two closely related species may be indistinguishable when analysed for a particular marker, leading to unreliable identifications. For example, conventional surveys detected the high-risk alien species Schizoporella japonica at seven of the nine stations investigated, but the species was not recorded from any eDNA samples. However, Schizoporella dunkeri was recorded in six of the nine stations investigated, and a detailed comparison of the marker region for these two species shows that they cannot be distinguished using the TAReuk marker, and the S. dunkeri occurrences in eDNA dataset likely represent misidentifications of S. japonica.
-
Mitigation measures : By compiling custom databases for use with probabilistic classification algorithms, databases can be quality-controlled to identify genera that experience poor marker performance and have a higher likelihood of identification errors.
4.1.3.2 - Database completeness
-
Large-scale simultaneous DNA-based species identification relies on comparisons to databases, which typically do not contain reference sequences from every single species that can possibly occur in the target area. As such, eDNA based methods will fail to correctly identify any species which does not appear in the reference database used for species identification. For example, conventional methods detect the annelid Spio martinensis , while eDNA methods do not. However, the NCBI GenBank database used in these analyses lacks a named COI reference sequence for this species, and as such it was not possible for the eDNA method to successfully detect it.
-
Mitigation measures : Taxonomists are continually generating new reference sequences and sharing them in public databases, allowing the DNA-based identification of an ever-broadening array of organisms. By regularly updating reference databases used for DNA-based identifications using public materials and collaborating with taxonomists to generate high-quality reference sequences for taxa previously not included in databases, we can continually improve the quality of DNA-based species identifications.
4.1.3.3 - Non-specific amplifications
-
To capture broad organismal groups, metabarcoding typically uses degenerate primers to capture the target region for the maximum number of species possible. However, amplification of random areas of a species’ genome can occur at a very low rate with these general primers. When employing best-match identification methods, such as the BLAST method used in this study, non-target fragments can nonetheless receive a species identification. However, these identifications are frequently incorrect, owing to the very small proportion of species having whole genome data available in public databases. As such these fragments are typically incorrectly identified as belonging to their nearest genome-sequenced relative. For example, the high-risk doorknocker species Ostrea chilensis was detected at two stations using eDNA metabarcoding, but further investigation of the sequences revealed them to be non-target amplifications that were most likely incorrectly identified.
-
Mitigation measures : DNA-based taxonomic identifications can be a time- and computationally intensive process. This pilot study employed a best-match taxonomic assignment method (BLAST) to identify sequences, which has the advantage of being quick and exploiting a large public database (NCBI GenBank). However, the method is highly sensitive to non-specific amplifications and database insufficiencies. Probabilistic taxonomic assignment methods (ex/ RDP classifier) will correctly discard non-target amplifications but are a trade-off because they have more demanding input requirements in the form of custom databases and training sets.
4.2 - eDNA metabarcoding as a tool for Horizon Scanning
Metabarcoding of settlement plates, zooplankton, sediment, and water samples using markers designed to capture broad organismal groups (Metazoa, Eukaryota) detected 2274 taxa of which 1640 could be identified to the species level and 681 of these were confirmed as native to Norway or as known alien/doorknocker species based on comparisons to the Norwegian Species Nomenclature Database and the Norwegian Alien Species List (Norwegian biodiversity and information centre).
However, 274 species (excluding micro-eukaryote, micro-algae, and microbes) were detected that could not clearly be attributed to known native or alien species in Norway. These taxa require expert evaluation to determine their status in Norway. Some may represent taxonomic nomenclature for native species that is inconsistent with the taxonomy used by the Norwegian Species Nomenclature Database and the Norwegian Alien Species List or species incorrectly identified due to the challenges described above.
However, others may be true occurrences of overlooked native species in Norwegian waters. For example, Acrochaetium catenulatum has not previously been recorded from the Norwegian coast but is known from Denmark and Sweden and likely represents a native species that has previously gone undetected. Finally, a subset of these species represents non-native species recently introduced to Norway. However, it must be stressed that species records belonging to this unclassified grouping should be interpreted with caution due to the challenges in DNA-based species identification discussed in Section 4.3.1. Nevertheless, these detections can be used to guide concerted, directed surveys to confirm the presence of newly introduced alien species, contributing to the early detection which is critical to the successful control and eradication of invasive species. Furthermore, these lists of species not currently known as native or alien in Norway can provide a data-driven knowledge base for horizon scanning and evaluation of their risk as doorknocker species.
4.3 - Recommendations for future monitoring of marine alien species
-
The number of sites in each port should follow the recommendations in the HELCOM protocol, which states that at least three sites should be investigated in each port and more sites should be examined in larger ports. There is currently no international recommendation regarding this, but we suggest that five to nine sites should be examined in the larger ports. Sites should be chosen based on a priori risk assessment of activity in the port that facilitates settlement of alien marine species, e.g., vessel activity at each quay or marina (Husa et al. 2023). Some areas in a port might be difficult to investigate for several days due to high activity and security concerns. LeClerck et al. 2018 found that alien marine species establish equally in large international ports and smaller ports nearby. This shows that choosing sites near the busiest areas also can be fruitful.
-
According to Hewitt et al. 2014 it is important to examine all available habitats in each port. This pilot study achieved to address all habitat types using settlement panels, investigations of marinas (RCS), dredge and video transects, traps, grab samples for infauna, zooplankton samples and beach surveys.
-
Based on the results from this pilot study we recommend that a combination of conventional methods and DNA methods should be used in a national monitoring program for marine alien species in Norway. Such a combination of methods has been successfully used in several countries. The conventional methods gave solid evidence of a species presence at the site, while the DNA analysis gave indications of multiple alien species that might be present but not found/identified with the conventional methods.
-
Rapid coastal survey (14) and settlement panels (10), followed by beach survey (9) yielded most alien species of the conventional methods in this study, whereas water samples (19), settlement plates (18) and zooplankton (14) found most alien species using DNA metabarcoding. RCS is relatively quick and detects many alien species and including more genetic samples of single specimen during the survey would increase the number alien species detected. For the collection of the settlement panels, we recommend that each plate is placed in a separate container and that the containing water is sieved to be able to catch motile fauna that will swim away from the plate. This was not done in the present study, and we did for example not find any Caprella mutica on the plates when examined in the lab.
-
The use of dredge and video transects recorded only seven alien species and it was time consuming to sort and identify as a lot of material was collected. This method was however the only conventional method that recorded the alien species Didemnum vexillum and Agarophyton vermiculophyllum . These two species are better monitored by using more targeted methods such as filming by underwater drone and the use of a grapnel in shallow areas for the latter. Beach surveys are important to include when possible because they will give a better picture of the abundance of molluscs and larger motile fauna such as prawns than the other conventional methods.
-
Crab pots and minnow traps did not detect a single alien species in this project, but we believe that in other localities, such as the Oslofjord for example, this method will provide important information on alien crabs. It is also a good method for catching benthic fish. The spread of alien Ponto-Caspian gobies are of concern and several species are now listed as doorknockers for Norway (Artsdatabanken 2023), including for example the round goby (Forsgren et al. 2023b). The fact that the traps caught ample numbers of native black gobies, a close relative, is promising. We recommend the use of minnow traps for sampling as the crab pots fished poorly.
-
Both conventional analysis of zooplankton and sediment samples yielded very few alien species, while DNA analysis of the same material recorded more. Identification of species by morphology is time consuming, requires extensive taxonomic skills and is often not possible for many cryptic taxa. We therefore suggest that sampling is made in the same way as in this study, but that half of the material goes to DNA analysis and the other half is fixated and stored till the metabarcoding results are ready and only examined for validation of species indicated in the DNA analysis. Zooplankton sampling can be improved by using passive plankton samplers over longer time intervals (e.g., three days at each site).
-
It would be a great advantage if metabarcoding results from waters samples were available before the start of the field work with conventional methods. This would give a more detailed target list for each station and improve the focus on special taxonomic groups and species. It would also be helpful to determine which species should be conserved for further genetic analysis. We therefore recommend that water samples for metabarcoding are taken in March/ April to secure that the results are ready before fieldwork in September the same year.
-
We also suggest that scraping samples for metabarcoding is taken from floating docks in marinas as well as from sediments on beaches .
-
The genetic analyses detected far more species in general, and more alien species than the conventional analyses. There is still large gaps and inconsistencies in the genetic reference databases, and some species are not possible to differentiate using our current genetic markers. Vice versa, there are many species that are difficult to differentiate using conventional methods that can be resolved using DNA analyses. Uncertainties in species identity in either method can often be resolved by using both methods.
-
DNA metabarcoding is a very general method and can discover new alien species (Couton 2022). However, monitoring single species can better be accomplished by single species analyses such as qPCR analyses. The use of species-specific qPCR for target listed species has been used in several studies (Andersen 2022). It might be a good idea also to develop a shorter target list of new arrivals which are expected to spread quickly for the Norwegian coast, like the ascidians Didemnum vexillum and Corella eumyota , the pacific shore crabs Hemigrapsus takanoi and H. sanguineus , the red algae Grateloupia turuturu , Ceramium sungminbooi and Agarophyton vermiculophyllum .
5 - References
Andersen, JH, Kallenbach E, Hesselsøe M, Knudsen, SW, Møller, PR, Bekkevold, D, Hansen, BK & Thaulow, J. 2016. Steps toward nation-wide monitoring of non-indigenous species in Danish marine waters under the Marine Strategy Framework Directive. Rapport 7022-2016 DK3. NIVA Denmark.
Andersen JH, Kallenbach E, Thaulow J, Hesselsøe M, Bekkevold D, Hansen BK, Jacobsen LMW, Olesen CA, Møller PR, Knudsen SW. 2018. Development of species-specific eDNA-based test systems for monitoring of non-indigenous species in Danish marine waters. RAPPORT L.NR. 7204-2017. Norwegian Institute for Water Research.
Andersen JH, Brink M, Kallenbach E, Hesselsøe M, Knudsen SW, Støttrup JG, Rask Møller P, Ekrem W, Fagerli C, Oug E. 2017. Sampling protocol for monitoring of non-indigenous species in selected Danish harbours. NIVA report 7175-2017.
Andersen, JH, Kallenbach, E, Kjeldgaard, MB, Knudsen, SW, Dale, T, Eikrem, W, Fagerli, C, Green, G, Hobæk, A, Oug, E, Thaulow, J, Hesselsøe, M, Bekkevold, D, Jacobsen, LMW, Kuhn, J, Møller, PR, Olesen, CA, Car, H & Stuer-Lauridsen, F. 2022. A baseline study of the occurrence of non-indigenous species in Danish harbours. NIVA Denmark Report 7769-2022.: 83.
Ardura, A, Zaiko, A, Martinez, JL, Samulioviene, A, Semenova, A & Garcia-Vazquez, E. 2015. eDNA and specific primers for early detection of invasive species – A case study on the bivalve Rangia cuneata , currently spreading in Europe. Marine Environmental Research 112, Part B: 48-55.
Ardura A, Zaiko A. 2018. PCR-based assay for Mya arenaria detection from marine environmental samples and tracking its invasion in coastal ecosystems. Journal for Nature Conservation 43: 1-7.
Artsdatabanken (2023). Fremmede arter i Norge – med økologisk risiko 2023. https://www.artsdatabanken.no/lister/fremmedartslista/2023
Berntsen HH, Sandlund OT, Thorstad EB, Fiske P. 2020. Pukkellaks i Norge, 2019. NINA Rapport 1821. Norsk institutt for naturforskning. https://hdl.handle.net/11250/2651741
Benjaminsen TM. 2022. Bryozoans in shallow water habitats in Norway: New records and DNA barcoding. Master’s thesis in Natural Science with Teacher Education. NTNU. 77pp.
Bringloe, T.T., Sjøtun, K. & Saunders, G.W. (2019). A DNA barcode survey of marine macroalgae from Bergen (Norway). Marine Biology Research https://doi.org/10.1080/17451000.2019.1699659
Couton, M, Comtet, T, Le Cam, S, Corre, E & Viard, F. 2019. Metabarcoding on planktonic larval stages: an efficient approach for detecting and investigating life cycle dynamics of benthic aliens. Management Of Biological Invasions 10(4): 657-689.
Couton M, Lévêque L, Daguin-Thiébaut C, Comtet T, Viard F. Water eDNA metabarcoding is effective in detecting non-native species in marinas, but detection errors still hinder its use for passive monitoring. Biofouling. 2022 Apr;38(4):367-383. doi: 10.1080/08927014.2022.2075739. Epub 2022 May 16. PMID: 35575060.
Dunshea G, Martell L, Bakken T, Budaeva N, Ekrem T, Tandberg AHS, Baussant T, de Boer H, Hestetun JT, Hobæk A, Kallioniemi EP, Larsen A, Markussen SS, Mauvusseau Q, Ray JL, Yoccoz and Willassen E 2021. Knowledge status for the use of molecular tools in mapping and monitoring biological diversity in marine environments. Miljødirektoratet Rapport M-2062.
Leclerc JC, Viard F, González Sepúlveda E, Díaz C, Neira Hinojosa J, Pérez Araneda K, Silva F, Brante A. Non-indigenous species contribute equally to biofouling communities in international vs local ports in the Biobío region, Chile. Biofouling. 2018 Aug;34(7):784-799. doi: 10.1080/08927014.2018.1502276. Epub 2018 Oct 24. PMID: 30354802.
Ekrem T, Baussant T, Dunshea G, Dunthorn M, Egge E, Engesmo A, Fossøy F, Hansen H, Kistenich S, Krolicka A, Larsen A, Mauvisseau Q, Strand DA, Vrålstad T & Westergaard KB 2023. The use of eDNA and DNA-based methods to assess and monitor alien and doorknocker species. Norwegian Environment Agency, Report M-2523. 63 pp.
Elbrecht, V, Braukmann, TWA, Ivanova, NV, Prosser, SWJ, Hajibabaei, M, Wright, M, Zakharov, EV, Hebert, PDN & Steinke, D. 2019. Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7: e7745.
Falkenhaug T, Agnalt A-L, Glenner H, Husa V, Jelmert A og Mortensen S (2023). Hoppekreps: Vurdering av Acartia tonsa for Fastlands-Norge med havområder. Fremmedartslista 2023. Artsdatabanken. https://www.artsdatabanken.no/lister/fremmedartslista/2023/1384
Fernandez, S, Miller, DL, Holman, LE, Gittenberger, A, Ardura, A, Rius, M & Mirimin, L. 2021. Environmental DNA sampling protocols for the surveillance of marine non-indigenous species in Irish coastal waters. Marine Pollution Bulletin 172: 112893.
Fossøy F, Sivertsgård R, Ambjørndalen VM, Brandsegg H, Andersskog IPØ, Husa V, Forsgren E. 2022. Kartlegging av den fremmede marine arten havnespy Didemnum vexillum ved hjelp av miljø-DNA. En rask respons undersøkelse. NINA Rapport 2092. Norsk institutt for naturforskning.
Fossøy, F., Husa, V, Andersskog, I.P.Ø., Brandsegg, H., Sivertsgård, R., Legrand, E., Svensen, Ø., Dahle Øverby, L., Husby E. 2023. Kartlegging av den fremmede marine arten havnespy Didemnum vexillum ved hjelp av miljø-DNA og visuell inspeksjon. Oppfølgende undersøkelser i 2022-23. NINA Rapport 2278. Norsk institutt for naturforskning.
Forsgren E, Bærum KM, Finstad AG, Gjelland KØ, Hesthagen T, Knutsen H, Wienerroither R. 2023a. Fisker: Vurdering av regnbueørret Oncorhynchus mykiss for Fastlands-Norge med havområder. Fremmedartslista 2023. Artsdatabanken. https://www.artsdatabanken.no/lister/fremmedartslista/2023/1762
Forsgren E, Bærum KM, Finstad AG, Gjelland KØ, Hesthagen T, Knutsen H, Wienerroither R. 2023b. Fisker: Vurdering av svartmunnet kutling Neogobius melanostomus for Fastlands-Norge med havområder. Fremmedartslista 2023. Artsdatabanken. https://www.artsdatabanken.no/lister/fremmedartslista/2023/1583
Forsström, T & Vasemägi, A. 2016. Can environmental DNA (eDNA) be used for detection and monitoring of introduced crab species in the Baltic Sea? Marine Pollution Bulletin 109(1): 350-355.
HELCOM 2013. HELCOM ALIENS 2- Non-native species port survey protocols, target species selection and risk assessment tools for the Baltic Sea. 34 pp.
Hewitt, CL, & Martin RB,2001 Revised Protocols for Baseline Port Surveys for introduced Marine Species: survey design, sampling protocol and specimen handling. http://www.cmar.csiro.au/e-print/open/CRIMP_Tech_Report_22.pdf
Hughey JR, Boo GH. (2016.). Genomic and phylogenetic analysis of Ceramium cimbricum (Ceramiales, Rhodophyta) from the Atlantic and Pacific Oceans supports the naming of a new invasive Pacific entity Ceramium sungminbooi sp. nov. Botanica Marina 59: 211-222.
Husa V, Agnalt A-L, Falkenhaug T, Fossøy F, Forsgren E, Grefsrud ES, Hansen F, Jelmert A, Mortensen S. 2022. Alien marine species in Norway. Mapping, monitoring and assessment of vectors for introductions. Rapport fra Havforskningen.
Husa V, Agnalt A-L, Falkenhaug T, Glenner H, Jelmert A og Mortensen S (2023). Mosdyr og begerormer: Vurdering av Juxtacribrilina mutabilis for Fastlands-Norge med havområder. Fremmedartslista 2023. Artsdatabanken. https://www.artsdatabanken.no/lister/fremmedartslista/2023/7340
Husa V, Heggøy E, Sjøtun K, Agnalt AL, Johansen PO, Glenner H, Hatlen K. 2012 a. Kartlegging av fremmede marine arter i Hordaland. Utredning for DN 2-2012. Direktoratet for naturforvaltning.
Husa V, Heggøy E, Agnalt A-L, Sjøtun K, Svensen R, Rokkan-Iversen K, Alvestad T. 2012 b. Kartlegging av fremmede marine arter i Rogaland. Utredning for DN 3-2012. Direktoratet for naturforvaltning.
Husa V, Agnalt AL, Svensen R, Rokkan Iversen K, Steen H, Jelmert A, Farestvedt E, Petersen H. 2013. Kartlegging av fremmede marine arter i indre og ytre Oslofjord. Utredning for DN 4-2013. Direktoratet for naturforvaltning.
Jensen T.C., Brandsegg H., Bækkelie K.A.E., Davey M., Fossøy F., Lungrin E., Majaneva M.; Schartau A.K., Velle G., Walseng B. 2021. DNA-baserte metoder som verktøy i overvåking av invertebrater i innsjøer. NINA Rapport 2044. Norsk institutt for naturforskning.
Leray, M, Yang, JY, Meyer, CP, Mills, SC, Agudelo, N, Ranwez, V, Boehm, JT & Machida, RJ. 2013. A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Frontiers in Zoology 10(1): 34.
Marraffini M, Ashton G, Brown C, Chang A, Ruiz G. 2017. Settlement plates as monitoring devices for non-indigenous species in marine fouling communities. MBI. 8:559–566. doi:https://doi.org/10.3391/mbi.2017.8.4.11
Minchin D. 2007. Rapid coastal survey for targeted alien species associated with floating pontoons in Ireland. Aquatic Invasions 2:63-70.
Miralles, L, Dopico, E, Devlo-Delva, F & Garcia-Vazquez, E. 2016. Controlling populations of invasive pygmy mussel ( Xenostrobus securis ) through citizen science and environmental DNA. Marine Pollution Bulletin 110(1): 127-132.
Mortensen S, Bodvin T, Strand Å, Holm MW, Dolmer P. 2017. Effects of a bio-invasion of the Pacific oyster, Crassostrea gigas (Thunberg, 1793) in five shallow water habitats in Scandinavia. Managing Biological Invasions 8: 543-552.
Moseid C., Falkenhaug T., Slettan A. 2021. Development of a TaqMan PCR assay for the identification of the non-native copepod Acartia tonsa, and detection of this species in Norwegian coastal waters, Journal of Plankton Research, Volume 43, Issue 3, May/June 2021, Pages 497–499, https://doi.org/10.1093/plankt/fbab029
Muñoz-Colmenero, M, Ardura, A, Clusa, L, Miralles, L, Gower, F, Zaiko, A & Garcia-Vazquez, E. 2018. New specific molecular marker detects Ficopomatus enigmaticus from water eDNA before positive results of conventional sampling. Journal for Nature Conservation 43: 173-178.
Polling, M., de Groot, G. A., Laros, I., & van den Heuvel-Greve, M. J. (2023). Identification of non-indigenous and other fouling species on marine litter using DNA metabarcoding: A comparison of marine taxa found on plastic litter and settlement plates collected from beaches and a harbour in the Netherlands, and a comparison to taxa found on marine litter in Iceland. (Report / Wageningen Environmental Research; No. 3253). Wageningen Environmental Research. https://doi.org/10.18174/629836
Radashevsky, V.I. Pseudopolydora (Annelida: Spionidae) from European and adjacent waters with a key to identification and description of a new species. Mar. Biodivers. 51, 31 (2021). https://doi.org/10.1007/s12526-020-01156-7
Rueness J, Heggøy E, Husa V, Sjøtun K (2007). First report of the Japanese alga Antithamnion nipponicum (Ceramiales, Rhodophyta) in Norway, and invasive species new to northern Europe. Aquatic Invasions 2: 431-434.
Savoie, A.M. & Saunders, G.W. (2015). Evidence for the introduction of the Asian red alga 1266 Neosiphonia japonica and its introgression with Neosiphonia harveyi (Ceramiales, Rhodophyta) 1267 in the Northwest Atlantic. Molecular Ecology 24: 5927-5937.
STOECK, T, BASS, D, NEBEL, M, CHRISTEN, R, JONES, MDM, BREINER, H-W & RICHARDS, TA. 2010. Multiple marker parallel tag environmental DNA sequencing reveals a highly complex eukaryotic community in marine anoxic water. Molecular Ecology 19(s1): 21-31.
Taberlet, P, Coissac, E, Hajibabaei, M & Rieseberg, LH. 2012. Environmental DNA. Molecular Ecology 21(8): 1789-1793.
Tryggestad, I. 2023. Assessment of a passive plankton sampler technology for monitoring densities of planktonic sea lice ( Lepeophtheirus salmonis & Caligus elongatus ) in the Hardangerfjord. MSc. Norwegian University of Science and Technology, Trondheim.
Van der Loos, Bafort Q, et al. 2023. Non-indigenous seaweeds in the Northeast Atlantic Ocean, the Mediterranean Sea and Macaronesia: a critical synthesis of diversity, spatial and temporal patterns. BioRxiv in press. doi: https://doi.org/10.1101/2023.06.05.543185
Zenetos, A, Tsiamis, K, Galanidi, M, Carvalho, N, Bartilotti, C, Canning-Clode, J, Castriota, L, Chainho, P, Comas-González, R, Costa, AC, Dragičević, B, Dulčić, J, Faasse, M, Florin, A-B, Gittenberger, A, Jakobsen, H, Jelmert, A, Kerckhof, F, Lehtiniemi, M, Livi, S, Lundgreen, K, Macic, V, Massé, C, Mavrič, B, Naddafi, R, Orlando-Bonaca, M, Petovic, S, Png-Gonzalez, L, Carbonell Quetglas, A, Ribeiro, RS, Cidade, T, Smolders, S, Stæhr, PAU, Viard, F & Outinen, O. 2022. Status and Trends in the Rate of Introduction of Marine Non-Indigenous Species in European Seas. Diversity 14(12): 1077.
6 - Appendix
6.1 - Appendix A - Available reference DNA sequences in international databases
Table 1. Algae: a list for marine alien algae species based on lists from Horizon scanning and risk assessment performed by the Norwegian Biodiversity Information Centre in 2022-2023. This target list contains alien species that are established in Norwegian waters, species that are observed here without documented viable populations and species that are considered door knockers. For each species, both Genbank and BOLD were investigated for the occurrence of reference DNA sequences.
| Species | Bold COI | Bold 28S | Bold rbcL | Genbank COI | Genbank rbcL | Genbank 28S | Genbank tufA |
| Botrytella parva | |||||||
| Fredericqia deveauniensis | X | X | X | X | X | X | |
| Gelidium vagum | X | X | X | X | X | ||
| Goniotrichopsis sublittoralis | |||||||
| Gracilaria vermiculophylla | X | X | X | X | X | X | |
| Grateloupia doryphora | X | X | X | ||||
| Grateloupia filicina | X | X | X | X | X | ||
| Grateloupia subpectinata | X | X | X | X | X | ||
| Grateloupia turuturu | X | X | X | X | X | X | |
| Halimeda incrassata | X | X | X | X | |||
| Kappaphycus alvarezii | X | X | X | X | X | ||
| Lomentaria hakodatensis | X | X | X | X | X | ||
| Gracilaria gracilis | X | X | X | X | X | X | |
| Myriactula areschougii | |||||||
| Myriactula clandestina | X | X | |||||
| Polyopes lancifolius | X | X | X | ||||
| Polysiphonia atlantica | X | X | X | ||||
| Polysiphonia morrowii | X | X | X | ||||
| Polysiphonia senticulosa | X | ||||||
| Sargassum muticum | X | X | X | X | X | X | |
| Solieria chordalis | X | X | X | ||||
| Ulva australis | X | X | X | X | X | ||
| Undaria pinnatifida | X | X | X | X | X | X | |
| Womersleyella setacea | X | X | X | ||||
| Bonnemaisonia hamifera | X | X | X | X | X | X | |
| Caulacanthus okamurae | X | X | X | ||||
| Colpomenia peregrina | X | X | X | X | |||
| Corynophlaea verruculiformis | |||||||
| Cryptonemia hibernica | |||||||
| Melanothamnus harveyi | X | X | X | X | X | ||
| Scytosiphon dotyi | X | X | |||||
| Sphaerococcus coronopifolius | X | X | X | X | X | X | |
| Ceramium sungminbooi | X | X | |||||
| Codium fragile | X | X | X | X | |||
| Codium parvulum | X | X | X | ||||
| Symphyocladiella dendroidea | X | X | X | X | X | ||
| Dasya baillouviana | X | X | X | ||||
| Dasya sessilis | X | X | X | X | |||
| Aegagropila linnaei | X | ||||||
| Agardhiella subulata | X | X | X | X | X | ||
| Aglaothamnion halliae | X | X | X | ||||
| Anotrichium furcellatum | X | X | |||||
| Antithamnion densum | X | X | |||||
| Antithamnion nipponicum | X | X | |||||
| Antithamnionella spirographidis | X | X | X | ||||
| Antithamnionella ternifolia | X | X | X | ||||
| Asparagopsis armata | X | X | X | X | X | X | |
| Dasysiphonia japonica | X | X | X | X | X | X | X |
| Ulvaria splendens | X | X | X | ||||
Table 2. Invertebrates: a list for marine alien invertebrate species based on lists from Horizon scanning and risk assessment performed by the Norwegian Biodiversity Information Centre in 2022-2023. This target list contains alien species that are established in Norwegian waters, species that are observed here without documented viable populations and species that are considered door knockers. For each species, both Genbank and BOLD were investigated for the occurrence of reference DNA sequences.
| Species | Bold COI | Genbank COI | Genbank 18S |
| Acartia tonsa | X | X | X |
| Alitta succinea | X | X | X |
| Alkmaria romijni | |||
| Ammothea hilgendorfi | X | X | X |
| Amphibalanus amphitrite | X | X | X |
| Amphibalanus improvisus | X | X | |
| Anadara inaequivalvis | X | X | |
| Aplidium glabrum | X | ||
| Arcuatula senhousia | X | X | X |
| Austrominius modestus | X | X | X |
| Beroe ovata | X | X | X |
| Bispira polyomma | |||
| Blackfordia virginica | X | X | X |
| Boccardia proboscidea | X | X | X |
| Boccardiella hamata | X | X | X |
| Botrylloides diegensis | X | X | X |
| Botrylloides violaceus | X | X | X |
| Bougainvillia rugosa | |||
| Brachidontes pharaonis | X | X | |
| Bugula neritina | X | X | X |
| Bugula stolonifera | X | X | X |
| Callinectes sapidus | X | X | X |
| Calyptospadix cerulea | |||
| Cancer irroratus | X | X | X |
| Caprella mutica | X | X | X |
| Celleporaria brunnea | X | X | |
| Celtodoryx ciocalyptoides | |||
| Cercopagis pengoi | X | X | X |
| Chama pacifica | X | ||
| Charybdis japonica | X | X | X |
| Chionoecetes opilio | X | X | X |
| Ciona robusta | X | X | |
| Ciona savignyi | X | X | X |
| Corambe obscura | X | X | X |
| Cordylophora caspia | X | X | X |
| Corella eumyota | X | X | X |
| Crassostrea gigas | X | X | |
| Crassostrea virginica | X | X | X |
| Crepidula fornicata | X | X | X |
| Crepidula onyx | X | X | |
| Cronius ruber | X | X | |
| Cryptorchestia cavimana | X | X | X |
| Desdemona ornata | |||
| Diadumene lineata | X | X | X |
| Didemnum vexillum | X | X | X |
| Edwardsiella lineata | X | ||
| Ensis directus | X | X | X |
| Eriocheir japonica | X | X | |
| Euspira catena | X | X | X |
| Evadne anonyx | |||
| Fenestrulina malusii | X | X | X |
| Ficopomatus enigmaticus | X | X | X |
| Gammarus tigrinus | X | X | X |
| Goniadella gracilis | |||
| Gonionemus vertens | X | X | |
| Grandidierella japonica | X | X | X |
| Haliclystus tenuis | X | X | |
| Heleobia australis | X | X | |
| Hemigrapsus sanguineus | X | X | |
| Hemigrapsus takanoi | X | X | X |
| Homarus americanus | X | X | X |
| Hydroides dianthus | X | X | |
| Hydroides elegans | X | X | X |
| Hydroides ezoensis | X | X | X |
| Indothais lacera | X | X | |
| Ischyrocerus commensalis | |||
| Jassa slatteryi | X | X | X |
| Limnoria quadripunctata | X | X | X |
| Limnoria tripunctata | |||
| Marenzelleria arctia | X | X | X |
| Marenzelleria neglecta | X | X | X |
| Marenzelleria viridis | X | X | X |
| Matuta victor | X | X | |
| Megabalanus coccopoma | X | X | X |
| Mercenaria mercenaria | X | X | X |
| Mnemiopsis leidyi | X | X | X |
| Molgula manhattensis | X | X | X |
| Monocorophium sextonae | X | X | X |
| Mulinia lateralis | X | X | X |
| Myicola ostreae | |||
| Mytilicola intestinalis | X | X | X |
| Mytilicola orientalis | X | X | X |
| Mytilopsis leucophaeta | |||
| Mytilopsis sallei | X | X | X |
| Mytilus chilensis | X | X | X |
| Naineris setosa | |||
| Nippoleucon hinumensis | |||
| Ocinebrellus inornatus | X | X | |
| Oculina patagonica | X | X | X |
| Oithona davisae | X | X | X |
| Ostrea chilensis | X | X | X |
| Pachycordyle navis | |||
| Pacificincola perforata | |||
| Palaemon macrodactylus | X | X | X |
| Paralithodes camtschaticus | X | X | X |
| Pectinatella magnifica | X | X | X |
| Penaeus aztecus | X | X | X |
| Penaeus japonicus | X | X | X |
| Penilia avirostris | X | X | X |
| Percnon gibbesi | X | X | |
| Perna viridis | X | X | X |
| Perophora japonica | X | X | X |
| Petricolaria pholadiformis | X | X | X |
| Phorcus sauciatus | X | X | |
| Pilumnus spinifer | X | X | |
| Pinctada imbricata | X | X | X |
| Polydora cornuta | X | X | X |
| Portunus segnis | X | X | |
| Potamocorbula amurensis | X | X | |
| Pseudodiaptomus marinus | X | X | X |
| Pseudopolydora paucibranchiata | X | X | X |
| Ptilohyale littoralis | |||
| Pyromaia tuberculata | X | X | |
| Rangia cuneata | X | X | X |
| Rapana venosa | X | X | X |
| Rhithropanopeus harrisii | X | X | X |
| Rhopilema nomadica | X | X | X |
| Ruditapes philippinarum | X | X | X |
| Schizoporella errata | X | X | |
| Schizoporella japonica | X | X | |
| Schizoporella unicornis | |||
| Spirobranchus kraussii | X | ||
| Streblospio benedicti | X | X | X |
| Styela clava | X | X | X |
| Symplegma reptans | X | X | X |
| Tapes philippinarum | X | X | |
| Teredo navalis | X | X | X |
| Theora lubrica | |||
| Tricellaria inopinata | X | X | |
| Urosalpinx cinerea | X | X | |
| Victorella pavida | |||
| Watersipora aterrima | |||
| Watersipora subtorquata | X | X | X |
| Xenostrobus securis | X | X | X |
Table 3 . Fish: a list for alien fish species that can occur in marine habitats based on lists from Horizon scanning and risk assessment performed by the Norwegian Biodiversity Information Centre in 2022-2023. This target list contains alien species that are established in Norwegian waters, species that are observed here without documented viable populations and species that are considered door knockers. For each species, both Genbank and BOLD were investigated for the occurrence of reference DNA sequences.
| Species | Bold COI | Bold 12S | Genbank COI | Genbank 12S |
| Acipenser oxyrinchus | X | X | X | X |
| Anguilla japonica | X | X | X | |
| Anguilla rostrata | X | X | X | X |
| Babka gymnotrachelus | X | X | X | |
| Coregonus muksun | X | X | ||
| Coregonus peled | X | X | X | |
| Fundulus heteroclitus | X | X | X | X |
| Morone americana | X | X | X | X |
| Neogobius fluviatilis | X | X | X | |
| Neogobius melanostomus | X | X | X | X |
| Oncorhynchus clarkii | X | X | X | |
| Oncorhynchus gorbuscha | X | X | X | X |
| Oncorhynchus keta | X | X | X | X |
| Oncorhynchus kisutch | X | X | X | X |
| Oncorhynchus mykiss | X | X | X | X |
| Oncorhynchus nerka | X | X | X | X |
| Oncorhynchus tshawytscha | X | X | X | X |
| Osmerus eperlanus | X | X | X | |
| Pelecus cultratus | X | X | X | |
| Ponticola kessleri | X | X | X | |
| Proterorhinus marmoratus | X | X | X | X |
| Salvelinus fontinalis | X | X | X | X |
| Vimba vimba | X | X | X |
6.2 - Appendix B - Salinity and temperature
Table 4 . Measurement of salinity and temperatures in the water column at each station. Values are given as mean values in the depth range of the measurement.
| Station | Date | Time | Depth | Salinity/psu | Temperature C ° |
| TAN1-CTD 1 | 29.08.2022 | 12:04 | 0-1 m | 31,5 | 16,6 |
| TAN1-CTD 1 | 29.08.2022 | 12:04 | 1-2 m | 31,8 | 15,4 |
| TAN1-CTD 1 | 29.08.2022 | 12:04 | 2-3 m | 32,1 | 16,3 |
| TAN1-CTD2 | 29.08.2022 | 12:15 | 0-1 m | 31,5 | 16,4 |
| TAN1-CTD2 | 29.08.2022 | 12:15 | 1-2m | 31,8 | 16,1 |
| TAN1-CTD2 | 29.08.2022 | 12:15 | 2-3 m | 32 | 15,8 |
| TAN1-CTD2 | 29.08.2022 | 12:15 | 3-4 m | 32,2 | 15,6 |
| TAN1-CTD 3 | 29.08.2022 | 12:19 | 0-1 m | 31,4 | 16,5 |
| TAN1-CTD 3 | 29.08.2022 | 12:19 | 1-2 m | 31,8 | 16,3 |
| TAN1-CTD 3 | 29.08.2022 | 12:19 | 2-3 m | 32 | 15,7 |
| TAN1-CTD 3 | 29.08.2022 | 12:19 | 3-4 m | 32,2 | 15,4 |
| TAN2-CTD 1 | 30.08.2022 | 13:09 | 0-2 m | 31,6 | 17 |
| TAN2-CTD 1 | 30.08.2022 | 13:09 | 1-2 m | 32 | 16,3 |
| TAN2-CTD2 | 30.08.2022 | 13:15 | 0-1 m | 31,6 | 17,1 |
| TAN2-CTD3 | 30.08.2022 | 13:19 | 0-1 m | 31,8 | 18,1 |
| TAN3-CTD 1 | 30.08.2022 | 13:50 | 0-1 m | 30,9 | 17,5 |
| TAN3-CTD 2 | 30.08.2022 | 13:59 | 0-1 m | 32,2 | 17,9 |
| EG1-CTD 1 | 31.08.2022 | 10:02 | 0-1 m | 6 | 17,5 |
| EG1-CTD 1 | 31.08.2022 | 10:02 | 1-2 m | 23,5 | 18,7 |
| EG1-CTD 1 | 31.08.2022 | 10:02 | 2-3 m | 25,9 | 18,6 |
| EG1-CTD 1 | 31.08.2022 | 10:02 | 3-4 m | 26,6 | 18,6 |
| EG1-CTD 1 | 31.08.2022 | 10:02 | 4-5 m | 26,7 | 18,4 |
| EG1-CTD 2 | 31.08.2022 | 10:47 | 0-1 m | 5,9 | 17,7 |
| EG1-CTD 2 | 31.08.2022 | 10:47 | 1-2 m | 20,6 | 18,1 |
| EG1-CTD 2 | 31.08.2022 | 10:47 | 2-3 m | 26,2 | 18,1 |
| EG1-CTD 2 | 31.08.2022 | 10:47 | 3-4 m | 27,4 | 17,8 |
| EG2-CTD 1 | 31.08.2022 | 11:59 | 0-1 m | 12,9 | 18,5 |
| EG2-CTD 1 | 31.08.2022 | 11:59 | 1-2 m | 24 | 18,8 |
| EG2-CTD 1 | 31.08.2022 | 11:59 | 2-3 m | 26 | 18,5 |
| EG2-CTD 1 | 31.08.2022 | 11:59 | 3-4 m | 26 | 18,5 |
| EG 3-CTD 1 | 31.08.2022 | 12:15 | 0-1 m | 13,7 | 18,5 |
| EG 3-CTD 1 | 31.08.2022 | 12:15 | 1-2 m | 24,3 | 18,8 |
| EG 3-CTD 1 | 31.08.2022 | 12:15 | 2-3 m | 25,6 | 18,7 |
| EG 3-CTD 2 | 31.08.2022 | 12:21 | 0-1 m | 13,2 | 18,4 |
| EG 3-CTD 2 | 31.08.2022 | 12:21 | 1-2 m | 24,9 | 18,8 |
| EG 3-CTD 2 | 31.08.2022 | 12:21 | 2-3 m | 22,9 | 18,9 |
| STA1-CTD 1 | 01.09.2022 | 08:36 | 0-1 m | 29,1 | 17,8 |
| STA1-CTD 1 | 01.09.2022 | 08:36 | 1-2 m | 29,2 | 17,7 |
| STA1-CTD 1 | 01.09.2022 | 08:36 | 2-3 m | 29,2 | 17,7 |
| STA1-CTD 2 | 01.09.2022 | 08:39 | 0-1 m | 29,1 | 17,8 |
| STA1-CTD 2 | 01.09.2022 | 08:39 | 1-2 m | 29,1 | 17,6 |
| STA1-CTD 3 | 01.09.2022 | 09:23 | 0-1 m | 29 | 17,9 |
| STA1-CTD 3 | 01.09.2022 | 09:23 | 1-2 m | 29 | 17,9 |
| STA1-CTD 3 | 01.09.2022 | 09:23 | 2-3 m | 29,1 | 17,8 |
| STA1-CTD 3 | 01.09.2022 | 09:23 | 8-9 m | 29,5 | 17,6 |
| STA2-CTD 1 | 01.09.2022 | 10:16 | 0-1 m | 29,2 | 17,9 |
| STA2-CTD 1 | 01.09.2022 | 10:16 | 1-2 m | 29,2 | 17,8 |
| STA2-CTD 1 | 01.09.2022 | 10:16 | 2-3 m | 29,2 | 17,8 |
| STA2-CTD 1 | 01.09.2022 | 10:16 | 3-4 m | 29,2 | 17,8 |
| STA2-CTD 2 | 01.09.2022 | 10:31 | 0-1 m | 29,2 | 17,8 |
| STA2-CTD 2 | 01.09.2022 | 10:31 | 1-2 m | 29,3 | 17,8 |
| STA2-CTD 3 | 01.09.2022 | 10:56 | 0-1 m | 29,5 | 17,6 |
| STA2-CTD 3 | 01.09.2022 | 10:56 | 1-2 m | 29,7 | 17,6 |
| STA2-CTD 3 | 01.09.2022 | 10:56 | 2-3 m | 29,7 | 17,5 |
| STA2-CTD 3 | 01.09.2022 | 10:56 | 3-4 m | 29,9 | 17,2 |
| STA3-CTD 1 | 01.09.2022 | 12:38 | 0-1 m | 28,8 | 18,4 |
| STA3-CTD 1 | 01.09.2022 | 12:38 | 1-2 m | 29 | 18,5 |
| STA3-CTD 1 | 01.09.2022 | 12:38 | 2-3 m | 29,4 | 17,7 |
| STA3-CTD 1 | 01.09.2022 | 12:38 | 5-6 m | 29,7 | 17,6 |
| STA3-CTD 2 | 01.09.2022 | 13:14 | 0-1 m | 28,8 | 18,3 |
| STA3-CTD 2 | 01.09.2022 | 13:14 | 1-2 m | 28,9 | 18,2 |
| STA3-CTD 2 | 01.09.2022 | 13:14 | 2-3 m | 29 | 18 |
| STA3-CTD 2 | 01.09.2022 | 13:14 | 5-6 m | 29,4 | 17,9 |
| STA3-CTD 3 | 01.09.2022 | 13:41 | 0-1 m | 28,7 | 18,4 |
| STA3-CTD 3 | 01.09.2022 | 13:41 | 1-2 m | 28,8 | 18,4 |
| STA3-CTD 3 | 01.09.2022 | 13:41 | 2-3 m | 28,8 | 18,4 |
| STA3-CTD 3 | 01.09.2022 | 13:41 | 5-6 m | 29,4 | 17,7 |
| STA3-CTD 3 | 01.09.2022 | 13:41 | 9-10 m | 29,7 | 17,6 |
6.3 - Appendix C – Potential marine alien species based on DNA metabarcoding.
Appendix Table 5. List of potential marine alien species that are not registered in the Norwegian species list, or in the Norwegian alien species list. These are potentially alien species for Norway but could also be native species that have not been recorded for Norway before. Species are assigned to the following tentative categories: D-LO: Doorknocker Low Risk, D-PH: Doorknocker Potential High Risk, O: possibly overlooked native, PC: possible contaminant, PI: possible introduction, PM: possible misidentification. Phyla are abbreviated to the following: AN=Annelida, AR=Arthropoda, BR= Bryozoa, CH=Chordata, CN=Cnidaria, CT=Ctenophora, MO=Mollusca, OC=Ochrophyta and RH=Rhodophyta.
|
Phylum |
Class |
Species |
Reported as alien in Europe (if not native) |
Nearest Occurrence |
Category |
|
AR |
Hexanauplia |
Acartia margalefi |
North Sea |
PM |
|
|
OC |
Phaeophyceae |
Acinetospora filamentosa |
North Sea/UK |
PI |
|
|
CN |
Anthozoa |
Alcyonium siderium |
N. America (East- coast) |
PI |
|
|
BR |
Gymnolaemata |
Alcyonidium verrilli |
N. America (east coast) |
PI |
|
|
RH |
Florideophyceae |
Botryocladia wrightii |
Yes |
UK |
PI |
|
RH |
Florideophyceae |
Antithamnionella spirographidis |
Yes |
UK |
D-PH |
|
RH |
Bangiophyceae |
Pyropia yezoensis |
Yes |
Baltic Sea/English Channel |
PI |
|
RH |
Florideophyceae |
Grateloupia subpectinata |
Yes |
English channel |
D-LO |
|
AR |
Hexanauplia |
Pseudocalanus mimus |
W Greenland, Bering Sea, NW Alaska |
PI |
|
|
AR |
Hexanauplia |
Paramphiascella fulvofasciata |
North Sea |
PI |
|
|
AR |
Malacostraca |
Solenocera crassicornis |
Yes |
E Mediterranean |
PI |
|
OC |
Phaeophyceae |
Cystoseira schiffneri |
Yes |
2 records in the Mediterranean |
PI |
|
AN |
Polychaeta |
Spirobranchus latiscapus |
Australia/NZ/Japan |
PI |
|
|
NE |
Chromadorea |
Chromadorita leuckarti |
Denmark |
O |
|
|
NE |
Chromadorea |
Punctodora ratzeburgensis |
Denmark |
O |
|
|
NE |
PalaeoNE |
Cephalothrix rufifrons; |
Sweden |
O |
|
|
AR |
Malacostraca |
Actaea racemosa |
NT |
||
|
MO |
Bivalvia |
Musculus lateralis |
N. America East Coast |
PI |
|
|
AN |
Polychaeta |
Pectinaria gouldii |
West Atlantic |
PI |
|
|
BR |
Gymnolaemata |
Bugulina fulva |
No |
North Sea |
PI |
|
MO |
Bivalvia |
Pandora neoaphidis |
NT |
||
|
MO |
Bivalvia |
Pandora nouryi |
UK |
NT |
|
|
OC |
Phaeophyceae |
Halothrix ambigua |
No |
Japan |
PI |
|
AR |
Hexanauplia |
Calanus euxinus |
Black Sea |
PI |
|
|
AR |
Ostracoda |
Aurila disparata |
PI |
||
|
OC |
Phaeophyceae |
Eudesme borealis |
No |
Japan |
PI |
|
RH |
Florideophyceae |
Antithamnion sparsum |
No |
Korea |
PI |
|
AN |
Polychaeta |
Polydora onagawaensis |
No |
Japan, Russia, N. America |
PI |
|
AR |
Arachnida |
Ameronothrus yoichi |
No |
Japan |
PI |
|
CH |
Actinopteri |
Ammodytes personatus |
No |
West US |
PC |
|
CH |
Actinopteri |
Chirolophis japonicus |
No |
Pacific |
PC |
|
NE |
PalaeoNE |
Cephalothrix hongkongiensis |
NE Pacific |
O |
|
|
OC |
Phaeophyceae |
Feldmannia mitchelliae |
Cosmopolitan species |
O |
|
|
RH |
Florideophyceae |
Entwisleia bella |
No |
Tasmania |
PI |
|
OC |
Phaeophyceae |
Laminarionema elsbetiae |
No |
Unknown |
O |
|
OC |
Phaeophyceae |
Ectocarpus crouaniorum |
Unknown |
O |
|
|
EC |
Echinoidea |
Lytechinus variegatus |
No |
Central America |
PI |
|
CN |
Hydrozoa |
Laomedea calceolifera |
North Sea |
O |
|
|
RH |
Florideophyceae |
Acrochaetium catenulatum |
One record in Sweden |
O |
|
|
AN |
Polychaeta |
Glycera americana |
No |
N America, Japan, Australia, NZ, |
PI |
|
AR |
Malacostraca |
Maja brachydactyla |
Atlantic, North Sea |
O |
|
|
AR |
Pycnogonida |
Pycnogonum tenue |
Yes |
Japan |
PI |
|
CN |
Staurozoa |
Manania uchidai |
NW Pacific and Arctic |
O |
|
|
CN |
Hydrozoa |
Bouillonactinia multigranosi |
No |
NE Pacific |
PI |
|
AR |
Hexanauplia |
Zygomolgus dentatus |
No |
Korea |
PI |
|
AR |
Hexanauplia |
Doropygus elegans |
No |
Japan |
PI |
|
AR |
Hexanauplia |
Critomolgus vicinus |
No |
Korea |
PI |
|
AR |
Hexanauplia |
Asterocheres aesthetes |
No |
Korea |
PI |
|
MO |
Gastropoda |
Rissoa auriscalpium |
Mediterranean |
PM |
|
|
NE |
Chromadorea |
Sabatieria mortenseni |
No |
New Zealand |
PI |
|
AR |
Hexanauplia |
Acartia hudsonica |
PM |
||
|
OC |
Phaeophyceae |
Silvetia compressa |
Baltic Sea/Iceland |
PI |
|
|
CH |
Chloropicophyceae |
Chloroparvula pacifica |
Recently described taxa, several records in Sweden |
O |
|
|
CH |
Pycnococcus provasolii |
O |
|||
|
OC |
Phaeophyceae |
Saccharina japonica |
Yes |
Japan |
PI |
|
RH |
Bangiophyceae |
Pyropia haitanensis |
Yes |
China |
N |
|
RH |
Florideophyceae |
Melanothamnus japonicus |
Yes |
NE Atlantic |
PI |
|
AN |
Polychaeta |
Sirsoe munki |
East Pacific |
PI |
|
|
RH |
Florideophyceae |
Palmaria decipiens |
No |
Sub-Antarctic, Antarctic |
PI |
|
PO |
Calcarea |
Sycettusa aff. hastifera OV-2012 |
E Africa |
PI |
|
|
CN |
Hydrozoa |
Turritopsis nutricula |
No |
West Atlantic |
PM |
|
OC |
Phaeophyceae |
Pylaiella washingtoniensis |
No |
Northwest Atlantic |
PM |
|
PO |
Calcarea |
Ute aff. syconoides OV-2012 |
Australia – New Zealand |
PI |
|
|
CH |
Ulvophyceae |
Ulva expansa |
No |
Northwest Atlantic, Indonesia |
PI |
|
NE |
Pilidiophora |
Hubrechtella juliae |
No |
Northwest Atlantic |
PI |
|
AN |
Polychaeta |
Protodrilus ciliatus |
France |
O |
|
|
AN |
Polychaeta |
Polycirrus carolinensis |
No |
North Amerika |
PI |
|
AN |
Polychaeta |
Polydora calcarea |
No |
Sweden |
O |
|
AR |
Hexanauplia |
Pseudonychocamptus spinifer |
No |
East Pacific |
PI |
|
AR |
Arachnida |
Acaromantis vespucioi |
No |
South America |
PI |
|
AR |
Hexanauplia |
Quinquelaophonte aurantius |
No |
New Zealand |
PI |
|
AR |
Hexanauplia |
Zaus unisetosus |
No |
Korea |
PI |
|
AR |
Ostracoda |
Hemicytherura kajiyamai |
No |
Sweden |
O |
|
AR |
Hexanauplia |
Herrmannella hoonsooi |
No |
Korea |
PI |
|
AR |
Hexanauplia |
Thompsonula hyaenae |
Sweden |
O |
|
|
AR |
Hexanauplia |
Synstellicola paracarens |
Mediterranean |
PI |
|
|
AR |
Malacostraca |
Acantholobulus bermudensis |
No |
Central and south Americas |
PI |
|
BR |
Gymnolaemata |
Fenestrulina thyreophora |
No |
Australia-New Zealand |
PI |
|
BR |
Gymnolaemata |
Escharoides angela |
No |
Pacific |
PI |
|
BR |
Stenolaemata |
Disporella pristis |
No |
Australia-New Zealand |
PI |
|
BR |
Gymnolaemata |
Bitectipora retepora |
No |
New Zealand |
PI |
|
CH |
Ulvophyceae |
Ulva partita |
No |
Japan, China |
PI |
|
CH |
Ulvophyceae |
Lychaete pellucidoidea |
No |
Eastcoast US |
PI |
|
CH |
Ulvophyceae |
Ulvella ramosa |
No |
US, Australia |
PI |
|
CH |
Chlorophyceae |
Pseudulvella consociata |
No |
US, Australia |
PI |
|
CH |
Chlorodendrophyceae |
Tetraselmis convolutae |
PI |
||
|
CH |
Chlorodendrophyceae |
Tetraselmis marina |
PI |
||
|
CH |
Pyramimonadophyceae |
Pyramimonas obovata |
PI |
||
|
CH |
Pyramimonadophyceae |
Pyramimonas australis |
Kattegat/Baltic Sea |
PI |
|
|
CH |
Chlorophyceae |
Microglena uva-maris |
Unknown |
PI |
|
|
CH |
Trebouxiophyceae |
Pumiliosphaera acidophila |
Italy |
PI |
|
|
CH |
Ulvophyceae |
Halochlorococcum porphyrae |
US |
PI |
|
|
CH |
Chlorodendrophyceae |
Tetraselmis astigmatica |
North America |
PI |
|
|
CH |
Pyramimonadophyceae |
Pyramimonas parkeae |
North America- Japan |
PI |
|
|
CH |
Ulvophyceae |
Blidingia dawsonii |
North America, North Sea |
PI |
|
|
CH |
Chlorophyceae |
Desmodesmus bicellularis |
PI |
||
|
CH |
Ulvophyceae |
Chaetomorpha spiralis |
NW Atlantic |
PI |
|
|
CH |
Chlorophyceae |
Chlamydomonas isabeliensis |
Canada |
PI |
|
|
CH |
Ascidiacea |
Phallusia ingeria |
North and south Atlantic |
PI |
|
|
CH |
Ascidiacea |
Corella inflata |
No |
NE Pacific |
PI |
|
CH |
Ascidiacea |
Pyura dura |
No |
Mediterranean, North Sea |
PI |
|
CH |
Actinopteri |
Acanthogobius hasta |
No |
NE pacific |
PI |
|
CH |
Actinopteri |
Platichthys stellatus |
No |
Pacific |
PI |
|
CH |
Ascidiacea |
Molgula provisionalis |
NE coast US |
PI |
|
|
CH |
Actinopteri |
Symphodus ocellatus |
Mediterranean |
PI |
|
|
CH |
Actinopteri |
Myzopsetta ferruginea |
NW Atlantic |
PI |
|
|
CH |
Actinopteri |
Zoarces elongatus |
NE Pacific |
PI |
|
|
CH |
Actinopteri |
Pleuronectes quadrituberculatus |
N-Pacific, Alaska |
PI |
|
|
CH |
Actinopteri |
Liparis agassizii |
Japan, Korea |
PI |
|
|
CH |
Actinopteri |
Glyptocephalus zachirus |
N-Pacific, Alaska |
PI |
|
|
CN |
Hydrozoa |
Amphinema dinema |
North Sea |
PI |
|
|
CN |
Staurozoa |
Calvadosia cruciformis |
No |
Japan |
PI |
|
CN |
Anthozoa |
Ostiactis pearseae |
US West coast |
PI |
|
|
CN |
Anthozoa |
Calcigorgia beringi |
Pacific |
PI |
|
|
CN |
Scyphozoa |
Cyanea tzetlinii |
White Sea |
PI |
|
|
CN |
Staurozoa |
Manania handi |
US, East coast |
PI |
|
|
CN |
Hydrozoa |
Garveia grisea |
Mediterranean |
PI |
|
|
CN |
Hydrozoa |
Rhacostoma atlanticum |
West Atlantic |
PI |
|
|
CN |
Hydrozoa |
Laomedea angulata |
North Sea |
PI |
|
|
CN |
Hydrozoa |
Hydra oligactis |
North Sea |
PI |
|
|
CN |
Hydrozoa |
Coryne uchidai |
Pacific |
PI |
|
|
CN |
Hydrozoa |
Hydra circumcincta |
North Sea |
PI |
|
|
CN |
Hydrozoa |
Stylactis fucicola |
Mediterranean |
PI |
|
|
CN |
Hydrozoa |
Coryne muscoides |
North Sea |
PI |
|
|
CN |
Anthozoa |
Actinia fragacea |
North Sea |
PI |
|
|
CN |
Hydrozoa |
Sertularia distans |
North Sea |
PI |
|
|
CN |
Hydrozoa |
Eudendrium carneum |
North Sea |
PI |
|
|
CN |
Anthozoa |
Actinia tenebrosa |
Australia/New Zealand |
PI |
|
|
CN |
Myxozoa |
Ellipsomyxa gobii |
PI |
||
|
CN |
Hydrozoa |
Stauridiosarsia cliffordi |
US West coast |
PI |
|
|
MO |
Bivalvia |
Modiolus barbatus |
Not recorded in Norway before, but natural migrant |
N |
|
|
MO |
Bivalvia |
Anadara broughtonii |
Japan, Gulf of Mexico |
PI |
|
|
MO |
Polyplacophora |
Plaxiphora albida |
Australia |
PI |
|
|
MO |
Bivalvia |
Vesicomya galatheae |
East Pacific |
PI |
|
|
MO |
Gastropoda |
Ercolania felina |
Australia-New Zealand |
PI |
|
|
MO |
Gastropoda |
Bulla vernicosa |
Pacific |
PI |
|
|
MO |
Gastropoda |
Doto eireana |
UK |
PI |
|
|
MO |
Gastropoda |
Pseudotalopia sakuraii |
West Pacific |
PI |
|
|
MO |
Gastropoda |
Wakauraia sakaguchii |
Japan |
PI |
|
|
MO |
Gastropoda |
Aplysia parvula |
France |
PI |
|
|
MO |
Bivalvia |
Magallana angulata |
Yes |
Netherlands, France |
PI |
|
MO |
Gastropoda |
Philine exigua |
Unknown |
PI |
|
|
MO |
Gastropoda |
Merelina tokunagai |
Unknown |
PI |
|
|
MO |
Gastropoda |
Boonea seminuda |
No |
US- west and east coast |
PI |
|
MO |
Bivalvia |
Gari maculosa |
No |
East Africa |
PI |
|
MO |
Bivalvia |
Galeomma turtoni |
France, UK |
PI |
|
|
NE |
Enoplea |
Thoracostoma trachygaster |
North East US |
PI |
|
|
NE |
Chromadorea |
Microlaimus tenuispiculum |
Kattegat, North Sea |
PI |
|
|
NE |
Chromadorea |
Paracyatholaimus intermedius |
Kattegat, North Sea |
PI |
|
|
NE |
Chromadorea |
Diplolaimella dievengatensis |
Kattegat, North Sea |
PI |
|
|
NE |
Chromadorea |
Daptonema hirsutum |
Kattegat, North Sea |
PI |
|
|
NE |
Chromadorea |
Panagrolaimus paetzoldi |
PI |
||
|
NE |
Chromadorea |
Prochromadorella antarctica |
Antarctic |
PI |
|
|
NM |
Pilidiophora |
Siphonenteron bilineatum |
Sweden |
O |
|
|
NM |
PalaeoNE |
Carinina ochracea |
Sweden |
O |
|
|
OC |
Phaeophyceae |
Microspongium stilophorae |
Kattegat |
PI |
|
|
OC |
Phaeophyceae |
Kuckuckia spinosa |
Sweden |
O |
|
|
OC |
Phaeophyceae |
Microspongium alariae |
UK |
PI |
|
|
OC |
Phaeophyceae |
Planosiphon filiformis |
UK, North sea |
PI |
|
|
OC |
Phaeophyceae |
Sargassum thunbergii |
yes |
Origin Japan, Korea Recorded in Sweden in 2019 |
PI |
|
OC |
Phaeophyceae |
Dictyota implexa |
Origin unknown, present in France, Spain, Netherlands |
PI |
|
|
OC |
Phaeophyceae |
Petalonia binghamiae |
US, Azores, Africa, Australia, New Zealand |
PI |
|
|
OC |
Phaeophyceae |
Himantothallus grandifolius |
Antarctica |
PI |
|
|
OC |
Phaeophyceae |
Adenocystis utricularis |
South America, New Zealand |
PI |
|
|
PO |
Homoscleromorpha |
Oscarella balibaloi |
Mediterranean |
PI |
|
|
PO |
Demospongiae |
Mycale (Carmia)sanguinea |
|
Mediterranean |
PI |
|
PO |
Demospongiae |
Hymeniacidon kitchingi |
UK |
PI |
|
|
PO |
Demospongiae |
Suberites pagurorum |
UK |
PI |
|
|
PO |
Calcarea |
Lelapiella incrustans |
No |
Africa |
PI |
|
PO |
Demospongiae |
Isodictya compressa;Isodictya frondosa |
No |
Africa |
PI |
|
PO |
Demospongiae |
Metschnikowia bicuspidata |
North Sea |
PI |
|
|
RH |
Florideophyceae |
Naccaria wiggii |
North Sea |
PI |
|
|
RH |
Florideophyceae |
Antithamnion decipiens |
Mediterranean, Azores |
PI |
|
|
RH |
Florideophyceae |
Antithamnion antillanum |
Mediterranean, Azores |
PI |
|
|
RH |
Florideophyceae |
Callithamnion collabens |
North and South Atlantic |
PI |
|
|
RH |
Florideophyceae |
Ceramium affine |
No |
Indian Ocean |
PI |
|
RH |
Florideophyceae |
Bonnemaisonia clavata |
No |
Mediterranean, UK |
PI |
|
RH |
Florideophyceae |
Hummbrella hydra |
No |
Australia, New Zealand, Chile |
PI |
|
RH |
Florideophyceae |
Lomentaria pinnata |
No |
Japan, Phillipines |
PI |
|
RH |
Florideophyceae |
Colaconema proskaueri |
No |
N America east |
PI |
|
RH |
Bangiophyceae |
Pyropia pseudolinearis |
No |
NE and NW Pacific |
PI |
|
RH |
Florideophyceae |
Dasya naccarioides |
Southwest Asia, New Zealand, Australia |
PI |
|
|
RH |
Florideophyceae |
Stenogramma interruptum |
No |
UK |
PI |
|
AR |
Temoridae |
Temora turbinata |
Baltic Sea |
PI |